3
Thermo Scientific Poster Note
•
PN-64086-ASMS-EN-0614S
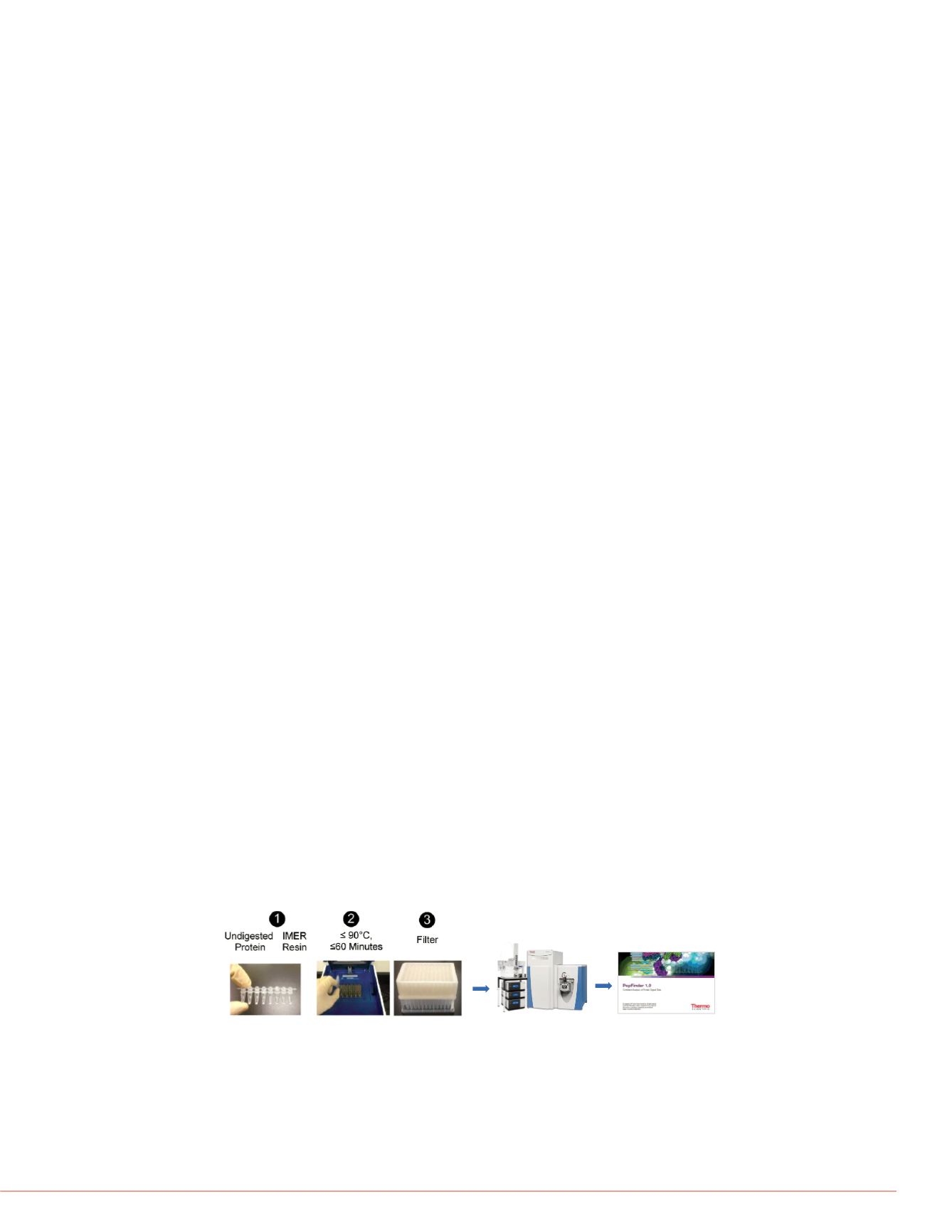
FIGURE1. Complete Workflow including Flash Digest, LC-MS and Data Analysis
Results
Tryptic Digestion Time Optimization of mAb by Flash Digest Kit
Flash Digest is a very active, highly stable immobilized trypsin reactor that is combined
with heating technology for fast reproducible digestions. The trypsin column makes
use of a high concentration of trypsin while simultaneously eliminating autolysis in
order to push the non-complete digestion due to the decrease of substrate
concentration near the completion of digestion reaction.
Using the Flash Digest kit (below workflow 1-3), digestion time was optimized by
incubating native, non-reduced IgG mAb at 70
°
C at 15, 30, 45, 60, 75, 90, 105 and
120 min. The filtered samples were directly subjected to LC-MS/MS and data analysis.
Sequence coverage maps of both li
mAb were generated from PepFinde
a 30-min digestion time is adequate
light chain and >79% for heavy chai
uncovered sequences on light (Figur
non-reduced disulfide bonds on cyst
coverage of light and heavy chains
120 min.
TABLE 1. Sequence Coverage Su
Digestion Times
Sequence
Coverage
15min 30min 45
Light Chain
78.5%
83.6%
83
Heavy Chain
79.1%
79.1%
79
sive characterization of
of post-translational
e enzymatically digested by
alyzed by online LC-MS on a
eptide sequence mapping,
by Thermo Scientific™
nd PTM analysis with a
combining rapid digestion,
der software. This workflow
is time while providing great
bs have been approved for
infectious and autoimmune
e quality of biotherapeutics
s have been used to study
, glycosylation pattern or
itive approach by combing fast
and user friendly new data
modifications analysis. This
in therapeutics in bioprocess
en peroxide and quenched by
ints. The native and
sing a Flash Digest kit
by incubating native, non-
to 120 minutes. One portion of
he other portion was reduced
purchased from Sigma Aldrich
were analyzed on Thermo
OAS
autosampler coupled to
separated on an ACQUITY®
column temperature set as 40
uoroacetic acid in H
2
O) and
Mass Spectrometry
The Q Exactive MS interfaced with H-ESI II ion source was employed for MS analysis.
Acquisition method was set with full scan (resolution 70,000 at FWHM
m/z
200) and
top 5 data dependent MS/MS (17,500 resolution) in positive mode.
Data Analysis
The mapping of mAb sequence, disulfide linkages and identification of PTMs are
performed in PepFinder software. PepFinder software is designed for in-depth
characterization of biotherapeutic proteins. It offers automatic workflow for
identification of disulfide bonds, glycopeptides and other PTMs, i.e. oxidation,
deamidation etc by mono-isotopic mass at MS level and confirmation by MS/MS
fragments indicated with a confidence score. The peptide sequence coverage map
with color code for signal intensity of each characterized peptide and modification
summary report with relative quantitation percentage are generated on the user
friendly interface. For unknown /untargeted modifications, the amino acid sites are
indicated with accurate mass of the modification for further interpretation.
=== HESI Source: ===
Spray Voltage (+) 3800V
Capillary Temperature (+) 320
°
C
Sheath Gas (+) 40
Aux Gas(+) 10
Sweep Gas(+) 0
Heater Temperature (+) 300
°
C
S-lens 50
Full MS Scan in positive mode: Resolution=70,000; AGC=3e6; IT=100ms;
Scan range=
m/z
300-1800; Lock mass=off; Microscans=1
Top 5 data dependent MS/MS: Resolution=17,500; AGC=1e5; IT=250ms;
NCE=27; Isolation window=
m/z
2; Fixed first mass=
m/z
130
LC-MS
Data Analysis
Relative
Abundance
15min
30min 4
N33+Deamidation
(Light Chain)
27.12%
28.12% 2
N162+Deamidatio
n (Light Chain)
15.98%
17.23% 1
M180+Oxidation
(Light Chain)
0.26%
0.35%
0
N83+Deamidation
(Heavy Chain)
1.44%
1.44%
1
TABLE 2. Selected PTMs of Nativ
FIGURE 2. Sequence Coverage M
Chain


