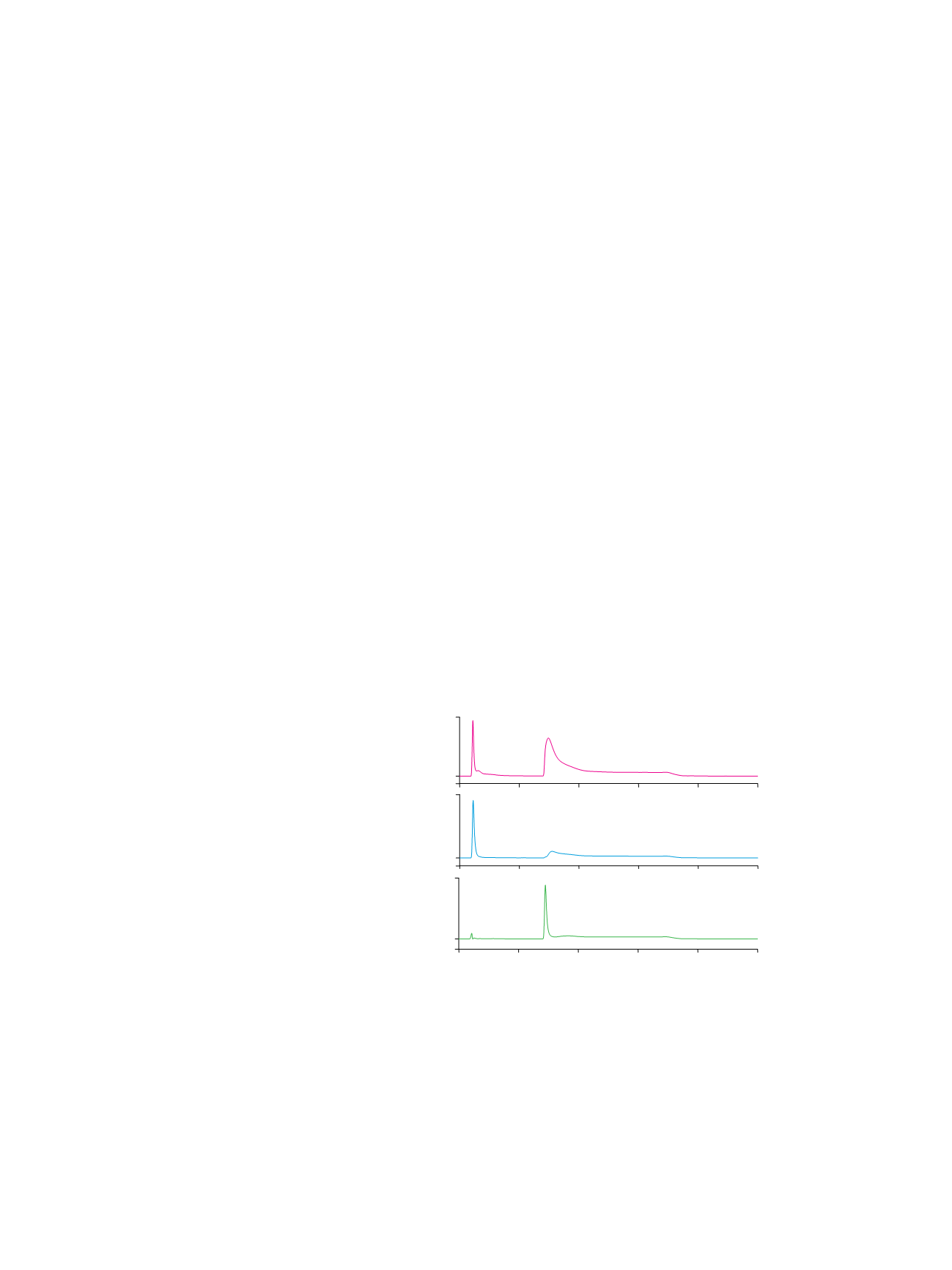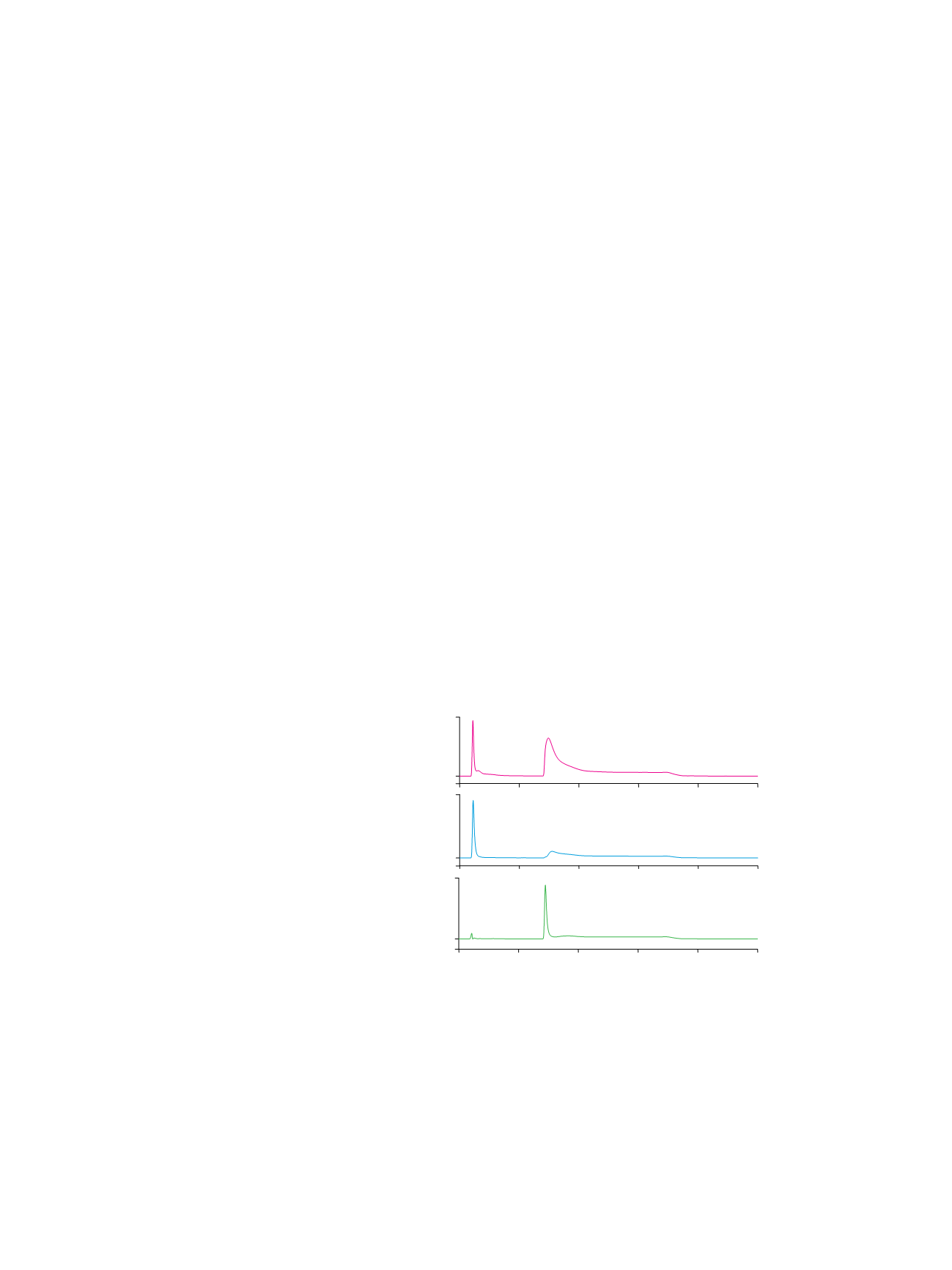
3
Preparation of Solutions
Buffers for Tryptic Digestion
For detailed methods for preparing buffers for tryptic
digestion, refer to Dionex (now part of Thermo Scientific)
Application Update (AU) 183.
1
Buffers for Deglycosylation
The buffers for deglycosylation are provided by New
England Biolabs, the Endo H manufacturer. The buffer
compositions are as follows: 10X Denaturation Buffer
[5% sodium dodecyl sulfate (SDS), 0.4 M dithiothreitol]
5X Reaction Buffer [0.5M sodium phosphate, pH 5.5].
Protein Digestion Procedure
Tryptic Digestion
Reduce, alkylate, and dialyze HRP extensively against
50 mM sodium bicarbonate. Digest the resulting HRP
with trypsin overnight. For detailed procedures, refer to
AU 183.
Deglycosylation
1. Add 40 µg of glycoprotein to an Eppendorf tube.
Prepare a 2 mg/mL solution by adding 20 µL DI water.
2. Add 2 µL 10X denaturation solution to the tube and
heat at 100 °C for 10 min.
3. Cool, then add 2 µL 5X reaction buffer to the tube.
4. Add 2 µL of Endo H to the reaction. Incubate overnight
at 37 °C.
Results and Discussion
Separation of a Glycoprotein from Its
Nonglycosylated Counterpart and/or
Nonglycosylated Impurities
Ovalbumin, ribonuclease B, and HRP are analyzed here
as glycoprotein models. Figure 1A shows that roughly
80% of the commercial ovalbumin can be captured by
the ProSwift ConA-1S Affinity column. A literature search
shows that ovalbumin has one
N
-linked glycosylation
site with approximately equal amounts of hybrid- and
high-mannose type oligosaccharides that can be recog-
nized by Con A. The other 20% of ovalbumin unbound
to Con A can likely be attributed to the contaminant
glycoproteins in ovalbumin that mainly have complex
type glycan structures.
2
Approximately 50% of ribonuclease B can be captured
by the ProSwift ConA-1S Affinity column (Figure 1B).
Ribonuclease B is reported to have a single glycosylation
site with high-mannose type oligosaccharide chains. The
ribonuclease B used in this study was labeled by the
manufacturer to be “a mixture of ribonuclease A and
ribonuclease B”. This result indicates that the commercial
ribonuclease B has roughly equal amounts of nongly-
cosylated ribonuclease A and glycosylated ribonuclease B.
As shown in Figure 1C, most if not all of the HRP was
captured by the ProSwift ConA-1S Affinity column.
This observation agrees with the fact that HRP has nine
potential glycosylation sites of which at least eight
sites are occupied by heterogeneous high-mannose
type oligosaccharides.
3
Interestingly, the eluted fraction (glycoprotein fraction)
peak of HRP was much sharper than the peak of
ovalbumin or ribonuclease B. This observation suggests
that HRP has a few dominant glycan structures with
similar affinities to Con A.
Figure 1. Glycosylated protein enrichment on the ProSwift ConA-1S Affinity column.
A
1
3
2
B
1
2
C
1
2
160
mAU
-20
-50
300
0
5
10
15
20
25
mAU
Minutes
400
-50
mAU
Column:
ProSwift ConA-1S Affinity (5 × 50 mm)
Mobile Phase:
A: 50 mM sodium acetate, 200 mM sodium chloride, 1 mM calcium
chloride, pH at 5.3
B: 100 mM
α
-methyl mannoside in mobile phase A
Gradient:
0–5.0 min, 0% B; 5.0–5.5 min,
0–100% B; 5.5–15 min, 100% B
Flow Rate:
0.5 mL/min
Inj. Volume:
20 µL
Temperature:
30 °C
Detection:
UV at 214 nm
Samples:
A
. Ovalbumin;
B
. Ribonuclease B;
C
. HRP
Sample Preparation: 1 mg/mL ovalbumin, ribonuclease B, or HRP in water/mobile phase A
Peaks:
1. Nonretained protein
2. Retained protein (nominally glycosylated)
3. Partially retained protein