6
Differentiate Minor Difference of Protein Structure in Biosimilar and Reference Products Using High-Resolution Orbitrap LC-MS/MSS
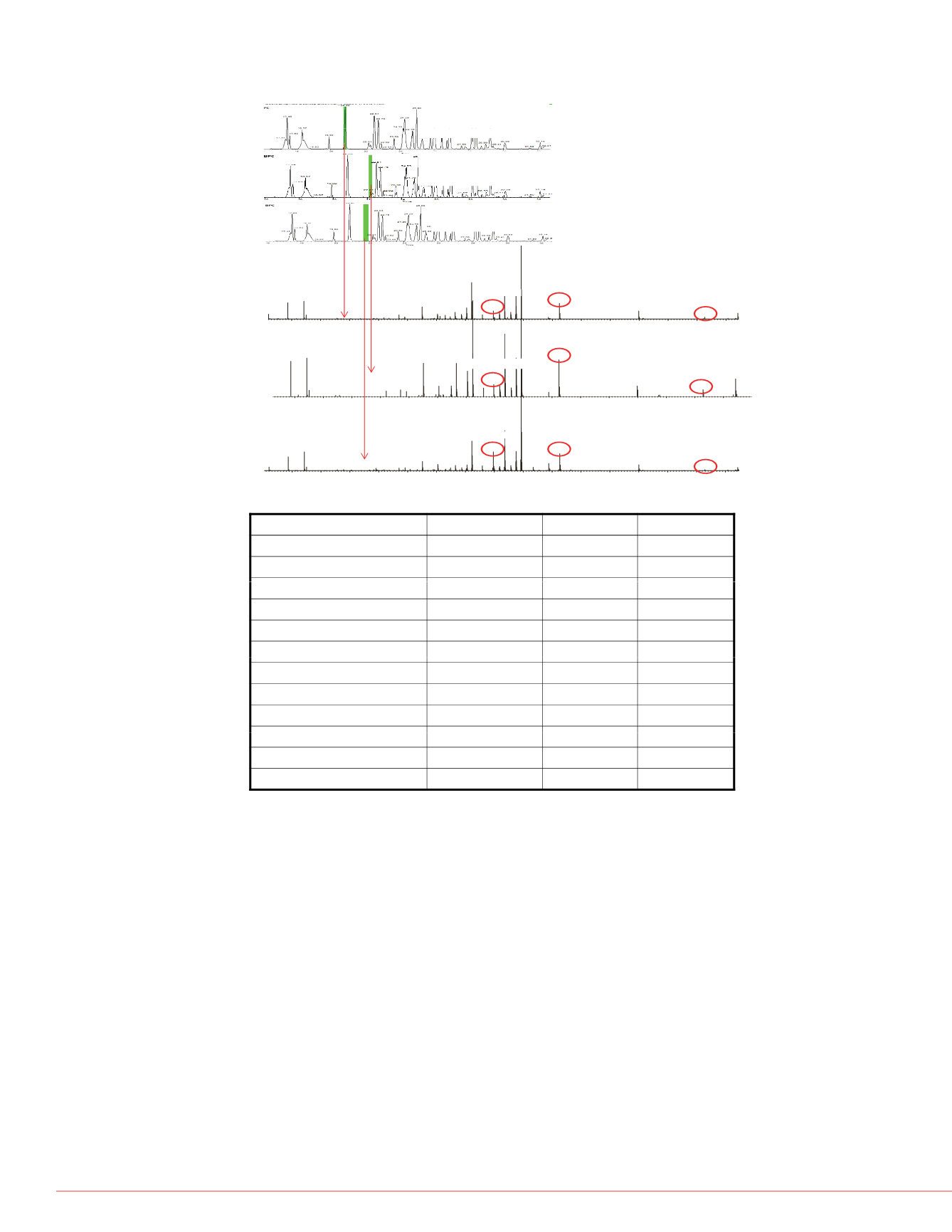
Figure 4. Identification and localization of two deamidation sites, N-140 and N-142 , on
peptide 136-LGLGNHNYCR-145. Base peak chromatogram (A) and high resolution HCD
spectra (B) of this peptide in native form or with deamidation either on N-140 or on N-142.
136-LGLGNHNY
C
R-145, 602.287 m/z
samples. Only those with relative
e samples are included.
a A, core fucose (Fuc) F, mannose
c acid (NANA) S, N-glycolyl
A
I-TNK
G-TNK
5.40%
3.23%
136-LGLG
N
HNY
C
R-145, 602.779, m/z, 12.24%
4607
y7++
517.2
y8++
NYCR
2.57%
ND
0.21%
1.79%
16.86%
14.43%
35 34%
37 59%
136-LGLGNH
N
Y
C
R-145, 602.779, m/z, 3.82%
no deamidation
b4++
b2
461.2
y7++
517.7
y8++
y9-H2O++
y4
546.2
y9++
150
200
250
300
350
400
450
500
550
600
650
700
750
800
850
900
m/z
110.1
143.1 171.1
b4++
b2
y1
335.1
y2
y8[3+]
375.2
y5++
401.9
M[3+]
432.2
y6++
452.2
y7-H2O++
.
479.2
498.2
y3
612.3
y4
749.3
y5
y6
920.4
y7
YCR
NYCR
NHNYCR
863.3
.
.
1.29%
0.80%
0.70%
0.56%
2.48%
0.91%
N 140 deamidation
B
150
200
250
300
350
400
450
500
550
600
650
700
750
800
850
900
m/z
143.1
171.1
y1
310.1 335.1
y2
y8[3+]
375.2
y5++
402.2
M[3+]
423.7
y6-H2O++
432.7
y6++
452.2
y7-H2O++
480.2
y3-H2O
498.2
y3
537.2
594.3
y4-H2O
612.3
749.3
y5
864.3
y6
921.4
y7
517.7
y8++
YCR
N
HNYCR
0.86%
0.57%
7.00%
5.04%
ND
2.20%
-
N 142 d id i
150
200
250
300
350
400
450
500
550
600
650
700
750
800
850
900
m/z
110.1
143.1
171.1
b4++
b2
y1
y2
345.5
y8[3+]
b7++
375.7
y5++
402.2
M[3+]
432.7
y6++
461.2
y7++
479.2
498.2
y3
528.7
537.7
y9-H2O++
567.2
594.3
613.2
y4
750.3
y5
921.4
y7
y6
YCR
N
YCR
NH
N
YCR
864.3
1.16%
0.56%
11.61%
16.50%
6.55%
2.62%
7 20%
6 51%
-
eam at on
Table 3. Identified deamidation sites and relative abundance of deamidation.
N117 Glycoform TPA
N117+A1G1M5
3 57%
.
.
C
Location of N-deamidation
TPA
I-TNK
G-TNK
N140
ND
12.24%
10.21%
N142
3.68%
3.82%
2.70%
.
N117+A1S1M4
2.63%
N117+A1S1M5
6.74%
N117+M5
52.41%
N205
2.08%
1.61%
0.15%
N218
0.63%
0.11%
0.31%
N234
0.15%
ND
ND
N37
29.83%
22.83%
19.64%
N117+M6
28.46%
N117+M7
6.00%
N 184 Glycoform I-TNK
D
N370
8.24%
13.56%
0.50%
N454
3.62%
2.71%
2.27%
N469
3.71%
2.05%
1.24%
N184+A2S1G1F
3.22%
N184+A2S2F
4.74%
N184+A3S2G1F
2.01%
N486
11.20%
10.80%
7.64%
N516
3.68%
2.87%
2.20%
N524
1.32%
0.51%
1.80%
Conclusion
An LC-MS/MS workflow was developed for differentiating minor difference of protein structure in
biosimilar and reference products. This workflow provides qualitative and quantitative comparison
of a biosimilar to a reference product.
N184+A3S3F
2.99%
N184+A4S3G1F
1.50%
N184+A4S4F
1.67%
1. 100% sequence coverage was obtained for all the nine data files analyzed. A five order
magnitude dynamic range for identified peptide abundance was achieved.
2. The identified covalent modifications, both expected and unexpected, include cysteine alkylation,
deamidation, overalkyation, Cys+DTT, oxidation, formylation, glycation. Relative abundance of
the modified forms was calculated and a comparison between files was generated.
uantified
that were indentified in these three
overalkyation, Cys+DTT, oxidation,
undance semi-tryptic and non tryptic
3. The site and type of glycosylation were identified and relative abundance of glycoforms was
calculated. Comparison of glycosylation sites, type and relative abundance of glycoforms
indicates the differences in glycosylation among the three samples.
entified in 3 different forms: native and
ely. A total of 12 deamidation sites were
s. Deamidation on N140 was only
es and relative abundance of N-
(T bl 3)
References
es a e .
All trademarks are the property of Thermo Fisher Scientific and its subsidiaries. This information is not intended to encourage use of
these products in any manners that might infringe the intellectual property rights of others.
1. Zhang Z, Shah B. Prediction of Collision-Induced Dissociation Spectra of Common N-
Glycopeptides for Glycoform Identification. Anal Chem 2010, 82: 10194–10202


