5
Thermo Scientific Poster Note
•
PN-64147-ASMS-EN-0614S
Top Down Analysis of Proteolytic Fragmented MAb
Middle down analysis of mAb protein was performed by LC-MS analysis of the
proteolytic fragments. Precursor ions at
m/z
964.65 of light chain,
m/z
1117.87 of Fd’ and
m/z
902.20 of the G0F glycoform of Fc/2 were selected respectively for ETD and HCD
0,000 resolution. (A) Full
chain; (C) Full MS spectrum
NL: 1.39E7
T: FTMS + p NSI Full
FIGURE 9. Comparison of exper
theoretically predicted isotope
and localizing G0F on Asn61 re
719 262
MS/MS at 120,000 resolution. Reaction time of 3-5 ms was applied for ETD
fragmentation. The normalized collision energy for HCD was 15-25 %. As shown in Figure
7, tandem spectra generated contain well resolved, multiply charged fragment ions.
Interpretation of these ions based on the mAb protein sequence was performed using
ProSightPC 3.0. The combined results of ETD and HCD suggest 50% sequence
ms [600.00-2000.00]
Fc/2
60
80
100
Abundance
.
z=9
719.1510
z 9
coverage for light chain, 52% coverage for Fc/2 with G0F and 32% coverage for Fd’
respectively (Figure 8, 10, 11). N-terminal modification of pyroglutamate of Fd’chain was
confirmed based on fragment ions from both ETD and HCD. Both intact and tandem
spectra identified the Lys loss at C-terminus of Fc/2.
ETD is widely known for its advantage in preferentially fragmenting the peptide back bone
NL: 2.67E7
T: FTMS + p NSI Full
ms [600.00-2000.00]
Light chain
0
20
40
Relative
=
719.0392
z=9
719 26
and keeping the labile modifications attached. It has been recognized as the method of
choice for locating the sites of such labile PTMs including glycosylation. Multiple identified
ETD fragments between Asn61 and Asn79 unambiguously located the glycan G0F on
Asn61 of Fc/2 chain. In the highly complex tandem spectrum, the high resolution and
accurate mass allows and is also necessary for confident identification of low abundance
1928.2022
z=12
NL: 1.75E7
60
80
100
.
z=9
719.1521
z=9
719.0407
glycan-containing fragment ions in the presence of interference. For example, shown in
Figure 7 insert is the identification of the +9 charged C
61
ion with G0F; Figure 9 presents
the unambiguous identification of C
57
without G0F. Although not all the isotopic peaks of
this c ion were observed due to background interference, all seven isotopes identified
were within 3 ppm mass error (external mass calibration). Both ETD ions, C
61
and C
57,
1976.9174
z=13
T: FTMS + p NSI Full
ms [600.00-2000.00]
Fd’
0
20
40
z=9
718.9293
z=9
have played critical roles in identification and localization of G0F on Asn61 residue.
FIGURE 10. ETD (blue) and HC
40 000 resolution (A)
2000
FIGURE 7. ETD spectrum of Fc/2 with G0F with precursor ion as
m/z
902.20,
charge +28. The insert is an expanded view of the spectrum covering C61 ion at
charge +9 with 3ppm mass accuracy, which confirms the addition of G0F
719.0
719.2
,
.
) Fd’, charge 21.
60
70
80
90
100
undance
939.2334
z=9
939.1216
z=9
939.3439
z=9
939.5667
z=9
90
100
764.9279
z=2
656 8907
+9, C
61
with
G0F, <3ppm
076
(A) F /2+G1F
glycan.
0
10
20
30
40
50
Relative Ab
70
80
nce
.
z=2
908 3
908 4
908 5
908 6
28
908.3156
z=28
c
FIGURE 11. ETD (blue) and H
939.0 939.2 939.4 939.6
m/z
40
50
60
elative Abunda
957.7858
z=3
1203.0937
z=4
924.4734
z=6 1073 0261
.
.
.
.
.4048
21
1102.4524
z=21
(B) Light chain
10
20
30
R
.
z=6
618.3590
z=1
358.2070
z=1
814.0227
z=5
1528.8462
z=1
1287.6293
z=5
4
1102.6
1102.8
1102.5968
z=21
1102.6899
z=21
200
400
600
800
1000
1200
1400
m/z
0
1224.4296
z=21
1224.5247
z=21
1224.6193
z=21
(C) Fd’
Conclusion
The middle-down study indicated t
224.4
1224.6
1224.8
ht chain.
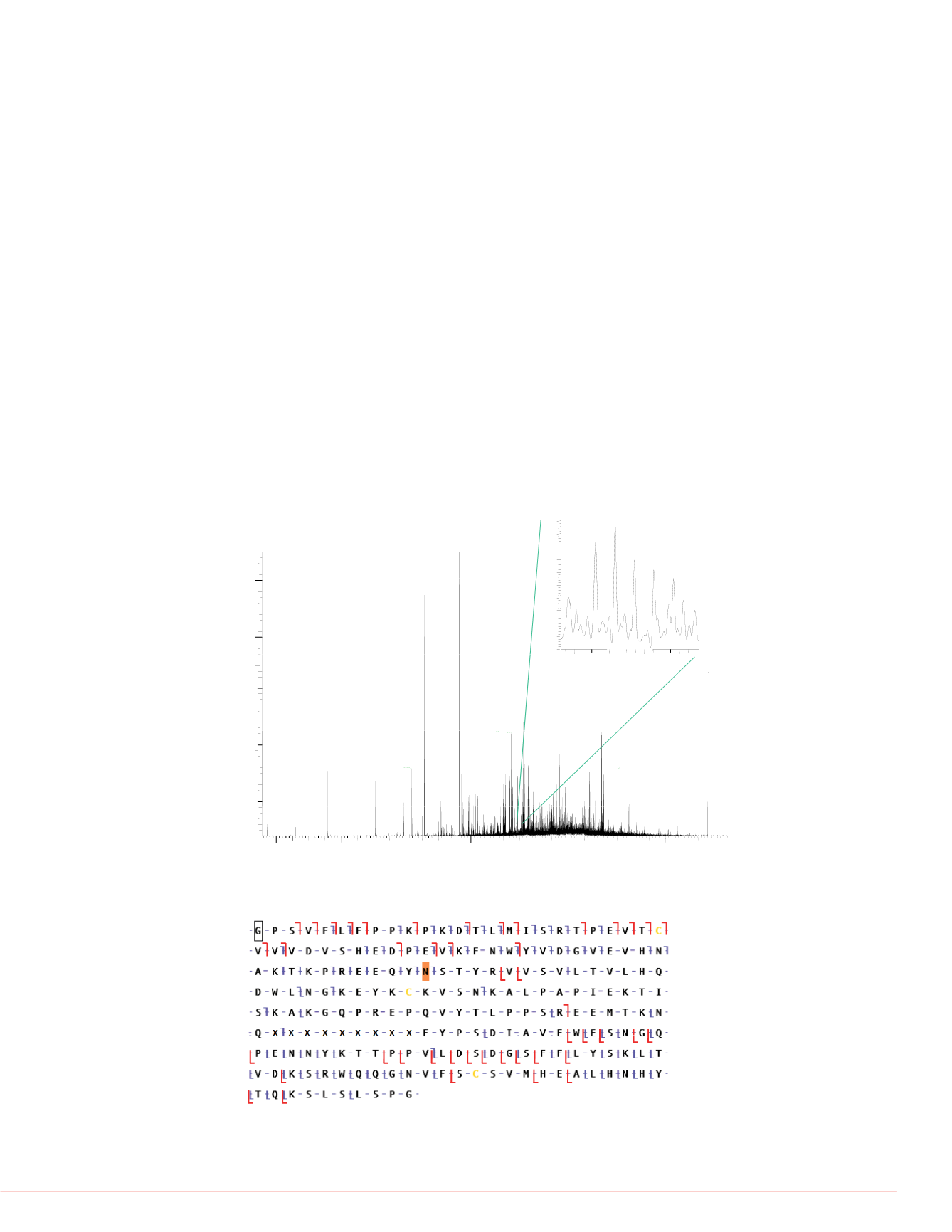
FIGURE 8. ETD (blue) and HCD (red) coverage of Fc/2+G0F chain. Asn61 was
highlighted for addition of G0F.
extensive sequence coverage as
including labile glycosylation. The
information deep into the middle o
Acknowledgeme
ppm
23113.3568
We would like to thank National In
intact mAb protein sample (candid
23100
23200
23130.3282
23096.2937
FabRICATOR® is a registered trademark for
Fisher Scientific and its subsidiaries. This inf
manners that might infringe the intellectual pr


