6
Structure Characterization and Differentiation of Biosimilar and Reference Products Using Unique Combination of Complementary Fragmentation Mechanisms
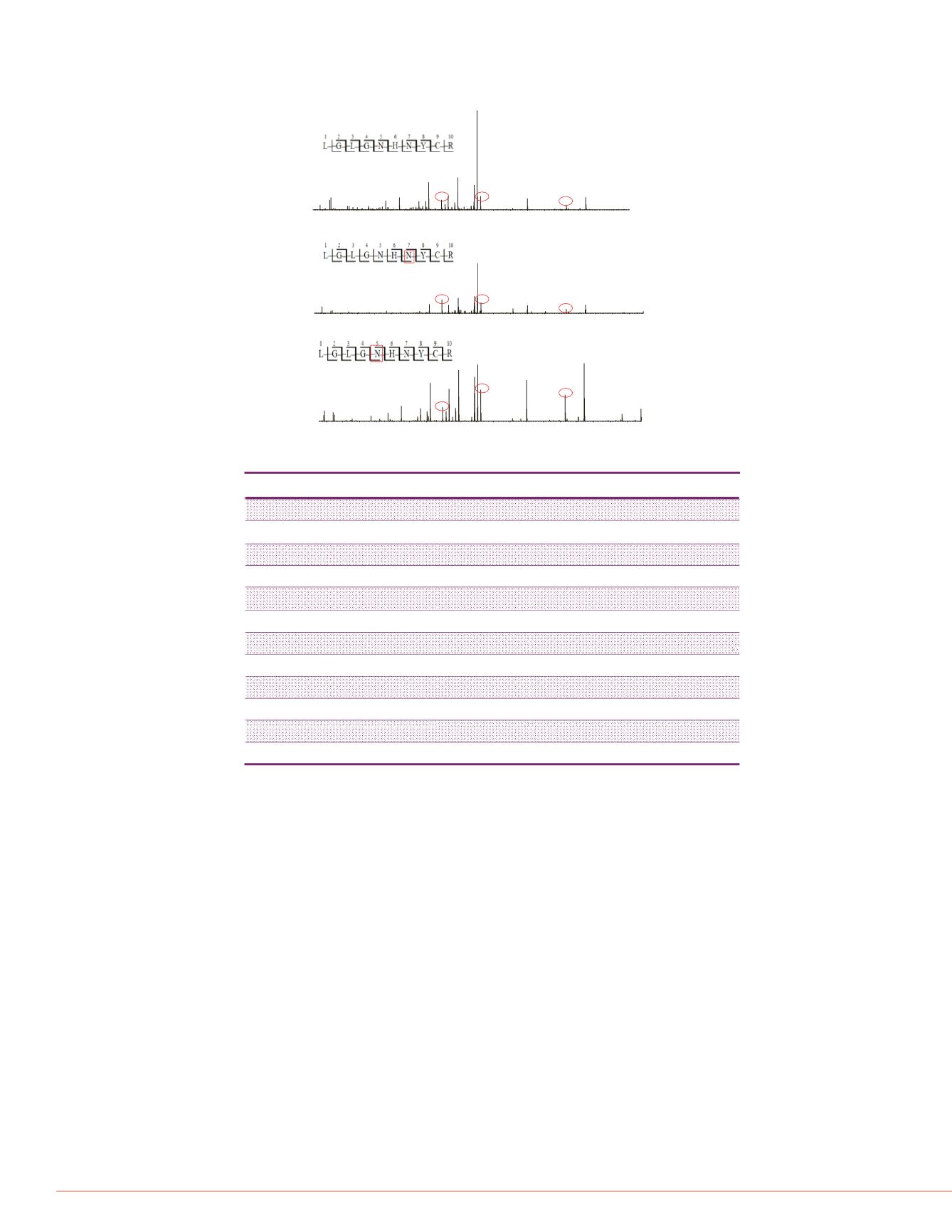
Figure 5. Identification and localization of two deamidation sites, N140 and N142 ,
on peptide L136-R145. High resolution HCD spectrum of this peptide in native form
(top), with deamidation either on N142 (middle) or on N140 (bottom).
were compared across the three
arious glycoforms on N448 were
602.3
M++
2). Most of glycans on this site
milar between I-TNK and G-TNK,
edly different. Although the most
two samples, the second and the
460.7
y7++
517.2
y8++
545.8
y9++
593.8
M-H2O++
y4
7
YCR
NYCR
NHNYCR
p five most abundant forms, only
(data not shown).
mannose, which is very different
sites (data not shown).
for the I-TNK sample (data not
602.8
M++
150
200
250
300
350
400
450
500
550
600
650
700
750
800
850
900
950
1000
m/z
143.1
b2
175.1
y1
223.1
284.2
b3
325.2
335.2
y2
b4
375.2
y5++
432.2
y6++
443.2
452.2
y7-H2O++
b5
498.2
y3
508.7
y8-H2O++
537.2
y9-H2O++
564.3
612.3
b7
749.3
y5
863.4
y6
b8
920.4
y
N142 deamidation.
Relative abundance = 10.99%
s that were indentified in these
idation, overalkyation, Cys+DTT,
acid.
150
200
250
300
350
400
450
500
550
600
650
700
750
800
850
900
950
1000
1050
m/z
145.1
b2
175.1
y1
335.2
y2
432.7
y6++
461.2
y7++
498.2
y3
517.7
y8++
546.2
y9++
b6
594.3
M-H2O++
613.2
y4
707.4
b7
750.3
y5
864.3
y6
b8
921.4
y7
y9
2.8
M++
YCR
N
YCR
NH
N
YCR
N140 deamidation.
ows confident identification and
W406. The relative abundance of
with high confidence in the three
150
200
250
300
350
400
450
500
550
600
650
700
750
800
850
900
950
1000
1050
145.1
171.1
b2
y1
228.1
284.2
b3
310.1
335.2
y2
b4
375.2
y5++
432.7
y6++
452.2
y7-H2O++
b5
461.2
y7++
498.2
y3
508.7
y8-H2O++
517.7
y8++
528.7
537.2
y9-H2O++
546.2
y9++
585.3
b6
593.8
M-H2O++
612.3
y4
707.4
b7
732.3
749.3
y5
864.3
y6
b8
903.3
y7-H2O
921.4
y7
b9
1034.5
y8
1091.5
y9
YCR
NYCR
N
HNYCR
Relative abundance = 5.93%
Table 3. Identified N-deamidation sites and relative abundance of deamidation
Location of N-deamidation
TPA
I-TNK
G-TNK
idi d tid T393 K416 d
d in I-TNK and G-TNK, but not in
across all three samples (Table 3).
entified in 3 different forms: native
ectively.
m/z
N140
ND
12.24%
10.21%
N142
3.68%
3.82%
2.70%
N205
2.08%
1.61%
<0.5%
ox ze pep e -
an
N218
0.63%
<0.5%
<0.5%
N234
<0.5%
<0.5%
<0.5%
N37
29.83%
22.83%
19.64%
N370
8.24%
13.56%
<0.5%
59.23
10937893
7871
N454
3.62%
2.71%
2.27%
N469
3.71%
2.05%
1.24%
N486
11.20%
10.80%
7.64%
.
.
376.2612
66.23
371.1023
89.77
371.103
CLPPADLQLPDWTECELSGYGK
Peak area = 1.98e+8
Conclusion
N516
3.68%
2.87%
2.20%
N524
1.32%
<0.5%
1.80%
60
65
70
75
80
85
59.83
17.4408
85.23
917.4407
64.41
917.4404
74.20
917.4401
A LC-MS/MS workflow was developed to differentiate minor differences in protein structure for
biosimilar and reference products using an Orbitrap Fusion instrument and new peptide mapping
software, PepFinder 1.0. This workflow provides qualitative and quantitative biosimilar to reference
product comparison.
1541.7
y13
1.
100% sequence coverage was obtained for all the nine data files analyzed .
2.
The identified covalent modifications, both expected and un-expected, include cysteine
alkylation, deamidation, overalkyation, Cys+DTT, oxidation, formylation, glycation and
glycosylation. The relative abundance of the modified forms was calculated and compared
between datasets. Confident identification and precise localization of low abundant PTMs was
1300
1400
1500
1600
1700
1800
1900
1329.6
y11
1444.6
y12
1654.8
y14
y15
achieved.
3.
Glycosylated peptides were characterized using the unique HCDpdETD method which generates
information of peptide sequence, site of glycosylation as well as glycan structure. Comparison of
glycosylation sites as well as the type and relative abundance of glycoforms indicated that there
are significant differences in glycosylation between the three samples.
1573.7
y13
nce= 0.09%
All other trademarks are the property of Thermo Fisher Scientific and its subsidiaries.
This information is not intended to encourage use of these products in any manners that might infringe the
intellectual property rights of others.
1361.6
y11
1476.6
y12
1686.7
y14
y15
PO64145-EN 0614S
1300
1400
1500
1600
1700
1800


