
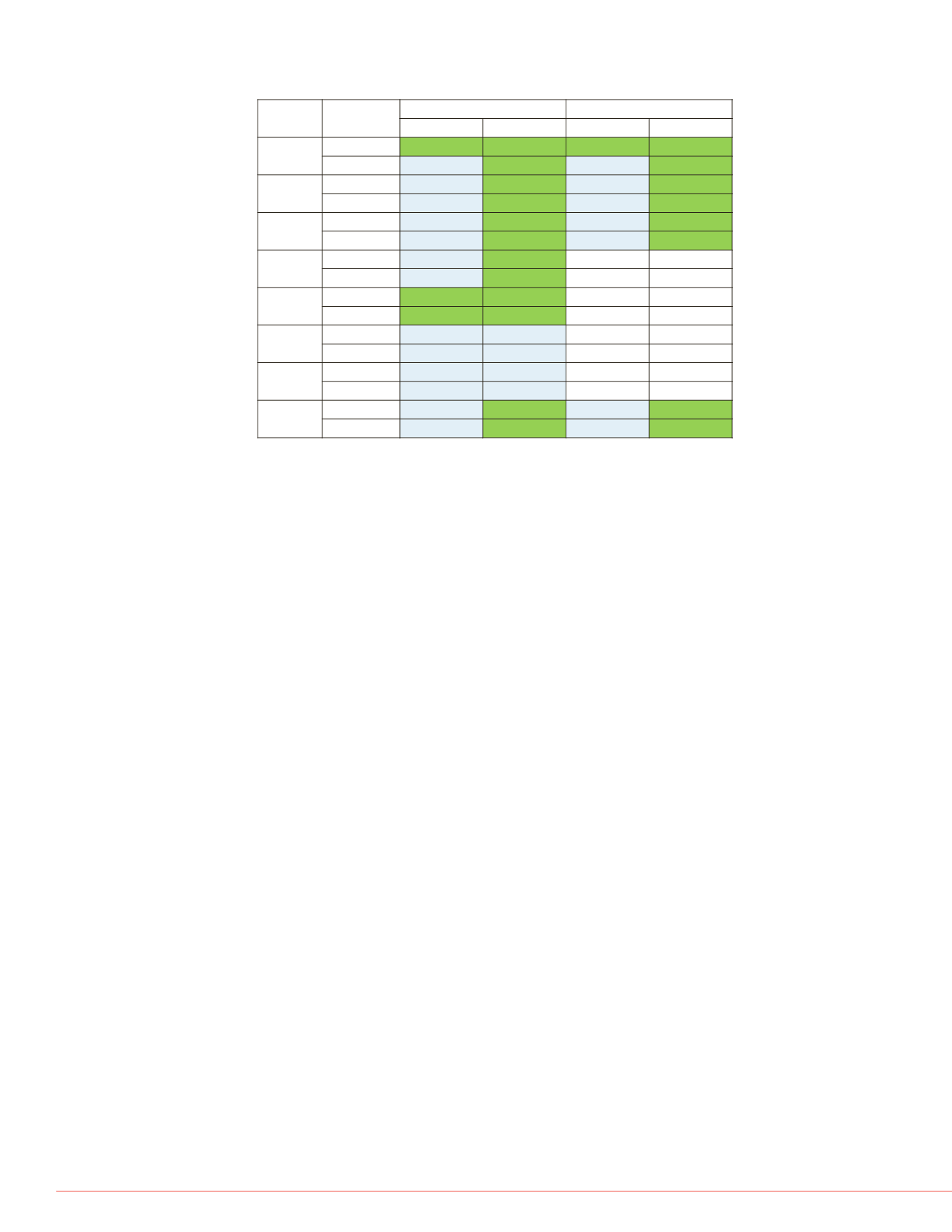
6
Enrichment of EGFR/PI3K/AKT/PTEN Proteins for Research using Immunoprecipitation and with Mass Spectrometry-based Analysis
GFR, AKT2, AKT1, PTEN, PIK3CA
ion. EGFR, AKT2, PTEN, PIK3CA
to 1000 fmol.
(fmol) ULOQ (fmol) Linearity (R
2
)
3.9
1000
0.9977
3.9
1000
0.9997
3.9
1000
0.9998
5.6
1000
0.9599
5.6
1000
0.9541
3.9
1000
0.9999
3.9
1000
0.9997
3.9
1000
0.9997
3.9
1000
0.9999
3.9
1000
0.9981
geted MS.
r quantitation of two unique EGFR
ved with Pierce Streptavidin (SA)
treptavidin T1 beads.
plication (nLC-SRM/MS)
ntitation of EGFR, AKT2, AKT1,
R) from plasma matrix.
ntitated at >7ng (52 fmol).
nLC-SRM/MS
FR Peptide Quantitation)
Streptavidin Magnetic Beads
ads
®
MyOne™ Streptavidin T1
293
Conclusion
Immunoprecipitation using magnetic beads for MS research applications
resulted in a higher yield of target protein and less non-specific binding than
using directly immobilized antibody.
Enrichment of EGFR, AKT isoforms, and PTEN in A431 and HEK293
lysates enabled detection by discovery MS and quantitation by targeted MS.
Immunoprecipitation of EGFR and AKT2 resulted in simultaneous analysis
of multiple isoforms and phosphorylation sites.
EGFR, AKT1, AKT2 and PTEN were quantified in the low nanogram range
by nLC-SRM/MS in two cell lysates.
Enrichment of as low as 7ng recombinant EGFR in plasma matrix and 10ng
of recombinant PIK3CA/PIK3R1 in cell lysate (data not shown) enabled
absolute quantitation by targeted SRM-MS.
Mulitplex IP to MS allowed simultaneous detection and quantification of
EGFR, AKT2, AKT1 and PTEN targets.
Future work will focus on optimization of IP conditions to enrich lower
abundant targets (<1ng total protein).
References
1. Logue JS, Morrison DK. Complexity in the signaling network: insights from the
use of targeted inhibitors in cancer therapy. Genes Dev. 2012 Apr 1; 26(7):641-
50.
2. Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes
using mass spectrometry. Nat Rev Mol Cell Biol. 2007 Aug; 8(8):645-54.
3. Carr SA, Abbatiello SE, Ackermann BL et al. Targeted Peptide Measurements in
Biology and Medicine: Best Practices for Mass Spectrometry-based Assay
Development Using a Fit-for-Purpose Approach. Mol Cell Proteomics. 2014 Mar;
13(3):907-17.
4. Ackermann BL . Understanding the role of immunoaffinity-based mass
spectrometry methods for clinical applications. Clin Chem. 2012 Dec;
58(12):1620-2.
Scaffold is a trademark of Proteome Software. All other trademarks are the property of Thermo Fisher Scientific
and its subsidiaries.
For Research use only. Not for use in diagnostic procedures.
This information is not intended to encourage use of these products in any manners that might infringe the
intellectual property rights of others.
FIGURE 9. Summary of EGFR-AKT pathway targets identified and quantified in
two cell lines without and with enrichment.
Target
Cell line
Detected by Orbitrap
Quantified by SRM
Neat
Enriched-IP
Neat
Enriched-IP
EGFR
A431
+
+
+
+
HEK293
-
+
-
+
AKT1
A431
-
+
-
+
HEK293
-
+
-
+
AKT2
A431
-
+
-
+
HEK293
-
+
-
+
AKT3
A431
-
+
N/A
N/A
HEK293
-
+
N/A
N/A
Grp94
A431
+
+
N/A
N/A
HEK293
+
+
N/A
N/A
PIK3CA
A431
-
-
N/A
N/A
HEK293
-
-
N/A
N/A
PIK3R1
A431
-
-
N/A
N/A
HEK293
-
-
N/A
N/A
PTEN
A431
-
+
-
+
HEK293
-
+
-
+
research applications.
ltaneously from HEK293 lysate with
vidin coated magnetic beads. All four
nLC-SRM/MS
e
Concentration
(fmol)
Concentration
(ng)
46
6.2
96
5.4
>ULOQ
>ULOQ
89
4.2
PO64139-EN 0614S



















