3
Thermo Scientific Poster Note
•
PN ASMS13_WP24_ELewis_e 06/13S
ted rapid trypsin digestion
C/MS/MS.
equipped trypsin column
ve proteins. This sample
rbitrap mass
lting data sets were
100% sequence coverage
chain were observed. For
ge of the light chain and
Furthermore, a digest of
he low coverage
in analytes can be
ered from the analytical
often the case that the
s delays the detection of
a lack of hands-free
results across multiple
ample preparation,
y alkylated monoclonal
erum albumin (1, 2).
ur at 60 C in 6 M
. Following reduction the
r at room temperature.
n in tris-buffered saline pH
rformed using a modified
with a Perfinity No
ers from Perfinity, a C18
(Halo 0.3 X 100 mm).
ystem. Digest efficiency
ous denaturation and
kylated samples were
r all samples.
microflow flow selector
ersed phase operations.
with the analytical column
volumes using initial
des were eluted with a 50
% formic acid in H
2
O
LC gradient was 0-50%B in
for automated peptide
Dionex™ nanoViper™
ted under isothermal
s utilized for all sample
rsed phase gradient. This
r sample is separated by
can be run every 7
a data-dependent top 10
uadrupole-Orbitrap mass
17.5K for HCD MS/MS
t parameters are listed in
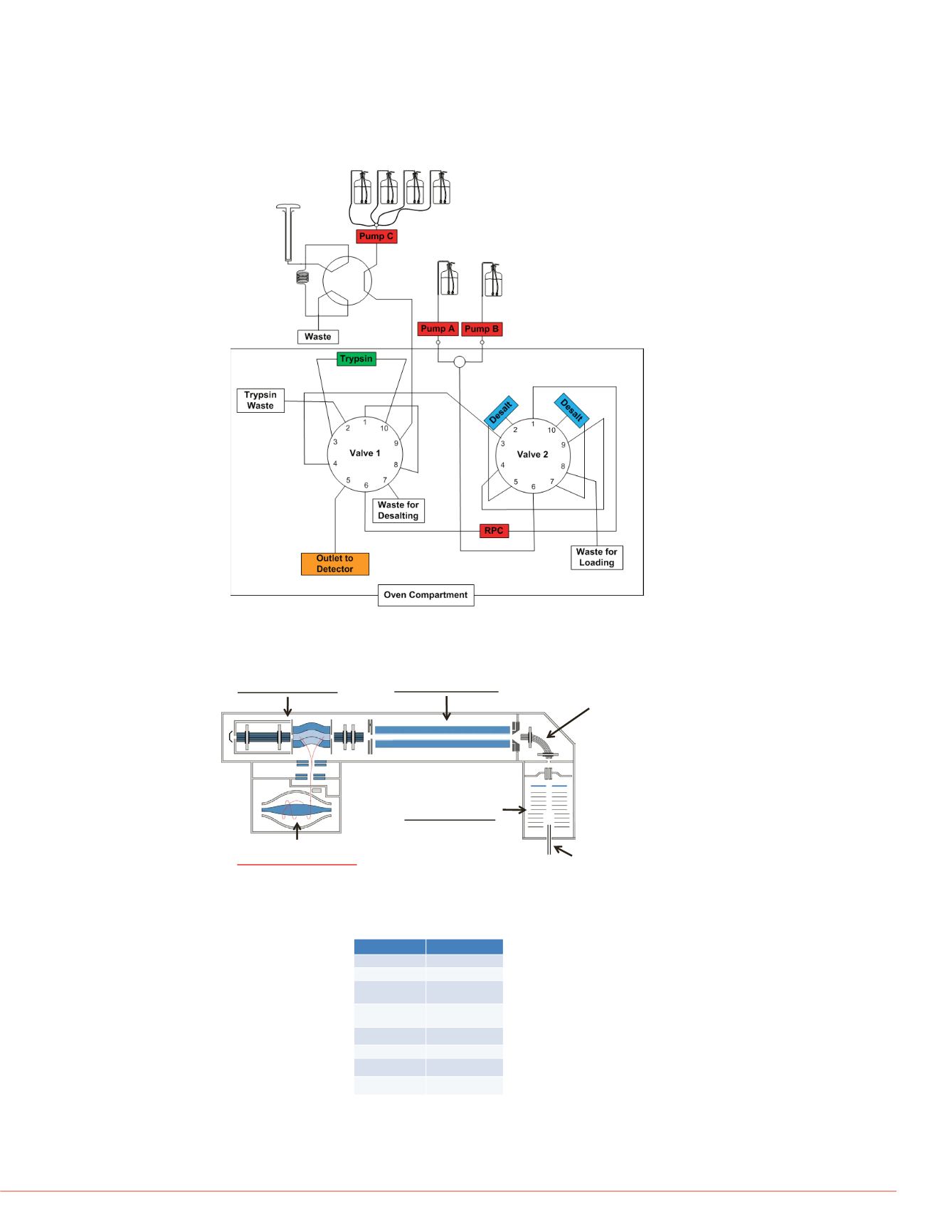
FIGURE 1. Schematics of the LC configuration for automated peptide mapping.
Results
Historically, Kumakura et. Al (2), showed t
was simply immobilized exhibited a 60% r
Shown in Figure 3, the proprietary stabiliz
dimensional structure enables prolonged
As shown in Figure 4, when the digestion
at various times and temperatures, a dra
70 C. These results suggest that under th
simultaneously denatured and enzymatic
results obtained by Vermeer et. al. that sh
variable regions occurs at 60 C and cons
FIGURE 4: Native mAb digested at vari
Figure 2. Schematic of the Q Exactive hybrid quadrupole-Orbitrap mass
spectrometer
.
Capillary
S-lens for
Improved Sensitivity
Bent
Flatapole
Quadrupole Mass Filter for
Precursor Ion Selection
HCD Cell, C-Trap for
Spectrum Multiplexing
Orbitrap with
Advanced Data Processing
Parameters
Settings
FullMS scan range
400-2000
MS/MSfixed firstmass
100
AGC
1e6, fullMS
5e4,MS/MS
Maxinjection time
(ms)
2, fullMS
60,MS/MS
Isolationwidth
(m/z)
2.0
NCE
27
Underfill ratio
1%
DynamicExclusion
30sec
Table 1. The Q Exactive hybrid quadrupole-Orbitrap mass spectrometer
instrument method parameters
Thermo Scientific
TM
Proteome Discoverer
TM
Software revision 1.3 was used to search the
protein database with the MASCOT
TM
search engine for all database searches. The
disulfide linked peptides were identified using StavroX (1). Raw files generated by the Q
Exactive hybrid quadrupole-Orbitrap mass spectrometer were searched directly using a
10 ppm precursor mass tolerance and a 20 amu fragment mass tolerance.
FIGURE 3. Data suggests that a trypsi
operation over prolonged periods at el


