3
Thermo Scientific Poster Note
•
PN-64155-ASMS-EN-0614S
on Orbitrap Elite.
yzed on LC-MS/MS tandem
digested samples, all with
lational modifications of
nd are crucial for their
fide bond match and mismatch
wing classes of
reatment of a variety of
, inflammation, and auto-
t heterogeneity, extensive
for a new mAb as a
essential tool in the
ybrid Ion Trap
-Orbitrap Mass
We analyzed the disulfide
es Rituxan® (Biogen
Roche) in Europe, is a
against the protein CD20. It
une therapy. Rituximab was
997 and by the European
phomas. The variable domain
that can be found in some non-
tal 16 disulfide bonds in
ide bond analysis, produces
ulfide bond analysis.
o different ways; one half was
was digested by chymotrypsin
ntific™
Accela HPLC system
1% formic acid in water,B:
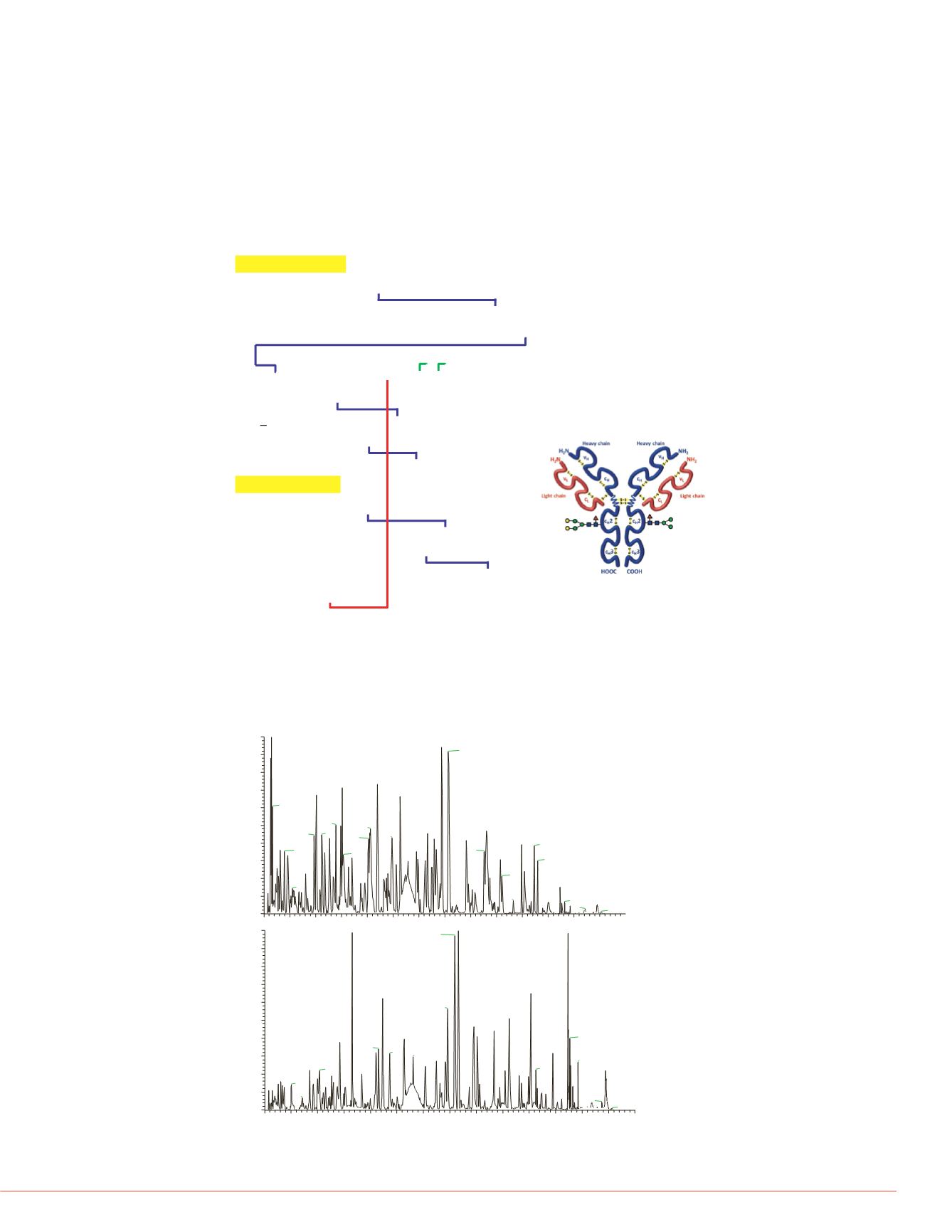
FIGURE 1. Amino acid sequence of heavy chain (top) and light chain (bottom)
Rituximab. The disulfide bonds (blue: intrachain linkage; red: interchain linkage
between light chain and heavy chain; green: interchain linkage between
different heavy chains) and glycosylation site on heavy chain (Asn residue
underlined and bolded) are also labeled.
Results
Overview
Rituximab is a recombinantly produced, monoclonal chimeric antibody against the
protein CD20, which has 16 disulfide bonds totally. FIGURE 1 shows the sequence
and disulfide bonds distribution of this mAb.
The result of trypsin-only digested
Using StavroX
TM
, we have identified
Rituximab. FIGURE 4 shows the su
μ
l/min
300
300
300
300
300
300
300
300
emperature of 275 °C.
ms; scan range: 300 to 2000 m/z;
00; TopN=12; NCE= 35.0
Goetze University of Halle-
Heavy Chain, length 451 aa
1
QVQLQQPGAE LVKPGASVKM SCKASGYTFT SYNMHWVKQT PGRGLEWIGA
51
IYPGNGDTSY NQKFKGKATL TADKSSSTAY MQLSSLTSED SAVYYCARST
101
YYGGDWYFNV WGAGTTVTVS AASTKGPSVF PLAPSSKSTS GGTAALGCLV
151
KDYFPEPVTV SWNSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ
201
TYICNVNHKP SNTKVDKKAE PKSCDKTHTC PPCPAPELLG GPSVFLFPPK
251
PKDTLMISRT PEVTCVVVDV SHEDPEVKFN WYVDGVEVHN AKTKPREEQY
301
N
STYRVVSVL TVLHQDWLNG KEYKCKVSNK ALPAPIEKTI SKAKGQPREP
351
QVYTLPPSRD ELTKNQVSLT CLVKGFYPSD IAVEWESNGQ PENNYKTTPP
401
VLDSDGSFFL YSKLTVDKSR WQQGNVFSCS VMHEALHNHY TQKSLSLSPGK
Light Chain, length 213 aa
1
QIVLSQSPAI LSASPGEKVT MTCRASSSVS YIHWFQQKPG SSPKPWIYAT
51
SNLASGVPVR FSGSGSGTSY SLTISRVEAE DAATYYCQQW TSNPPTFGGG
101
TKLEIKRTVA APSVFIFPPS DEQLKSGTAS VVCLLNNFYP REAKVQWKVD
151
NALQSGNSQE SVTEQDSKDS TYSLSSTLTL SKADYEKHKV YACEVTHQGL
201
SSPVTKSFNR GEC
FIGURE 2 shows the chromatogram of the trypsin-only digested and chymotrypsin-
trypsin double digested samples, and FIGURE 3 shows the sequence coverage of
trypsin-only digested and chymotrypsin-trypsin double digested samples, using the
software Proteome Discoverer 1.4. It can be found that both of the coverage rates are
nearly 100%, suggested the digestions went to completion.
FIGURE 3. the sequence converge m
FIGURE 4. All of the identified dis
Rituximab.
RT:
0.00 -70.00
0
5
10
15
20
25
30
35
40
45
50
55
60
65
Time (min)
0
10
20
30
40
50
60
70
80
90
100
RelativeAbundance
1.45
34.38
35.65
21.94
15.14
10.12
26.34
1.63
13.92
20.63
43.07
31.68
11.16
9.71
24.79
20.26
39.15
49.86 52.33
3.13 3.93
42.66
29.48
15.33
45.73
52.97
8.05
46.05
57.31
5.50
48.22
55.02 58.19 64.49
62.14 65.27
NL:
1.66E7
BasePeakF:
FTMS+pESI
Fullms
[300.00-
2000.00] MS
2s-chymotry
RT:
0.0 -70.00
0
5
10
15
20
25
30
35
40
45
50
55
60
65
70
Time (min)
0
10
20
30
40
50
60
70
80
90
100
RelativeAbundance
36.58
16.53
57.32
35.94
50.28
22.27
34.63
46.21
39.53 43.36
57.67
26.36
14.23
21.52
54.45
23.66 28.06 32.46
59.23
51.27
8.50
64.42
10.39
18.33
48.09
3.06 5.05
7.11
63.71
65.68
NL:
7.38E6
BasePeakF:
FTMS+pESI
Fullms
[300.00-
2000.00] MS
2s-try
Chymotrypsin+trypsin
Trypsin only
FIGURE 2. the chromatogram of the two samples.
FIGURE 5. The annotated MS/MS
heavy chain (trypsin digested on


