4
Disulfide Bond Analysis on Orbitrap Mass Spectrometry
in (top) and light chain (bottom)
ain linkage; red: interchain linkage
nterchain linkage between
on heavy chain (Asn residue
al chimeric antibody against the
y. FIGURE 1 shows the sequence
FIGURE 7. The annotated
chain (double digestion).
The result of trypsin-only digested sample
Using StavroX
TM
, we have identified 10 disulfide bonds in the trypsin-only digested
Rituximab. FIGURE 4 shows the summary of all identified disulfide bonds.
GRGLEWIGA
CARST
TAALGCLV
SSLGTQ
VFLFPPK
KTKPREEQY
GQPREP
NYKTTPP
KSLSLSPGK
IYAT
PTFGGG
QWKVD
VTHQGL
n-only digested and chymotrypsin-
shows the sequence coverage of
uble digested samples, using the
d that both of the coverage rates are
ompletion.
FIGURE 3. the sequence converge map of mAb digested in two different ways.
FIGURE 4. All of the identified disulfide linkage in trypsin-only digested
Rituximab.
45
50
55
60
65
49.86 52.33
45.73
52.97
46.05
57.31
48.22
55.02 58.19 64.49
62.14 65.27
NL:
1.66E7
BasePeakF:
FTMS+pESI
Fullms
[300.00-
2000.00] MS
2s-chymotry
45
50
55
60
65
70
57.32
50.28
46.21
.36
57.67
54.45
59.23
51.27
64.42
48.09
63.71
65.68
NL:
7.38E6
BasePeakF:
FTMS+pESI
Fullms
[300.00-
2000.00] MS
2s-try
Chymotrypsin+trypsin
Trypsin only
les.
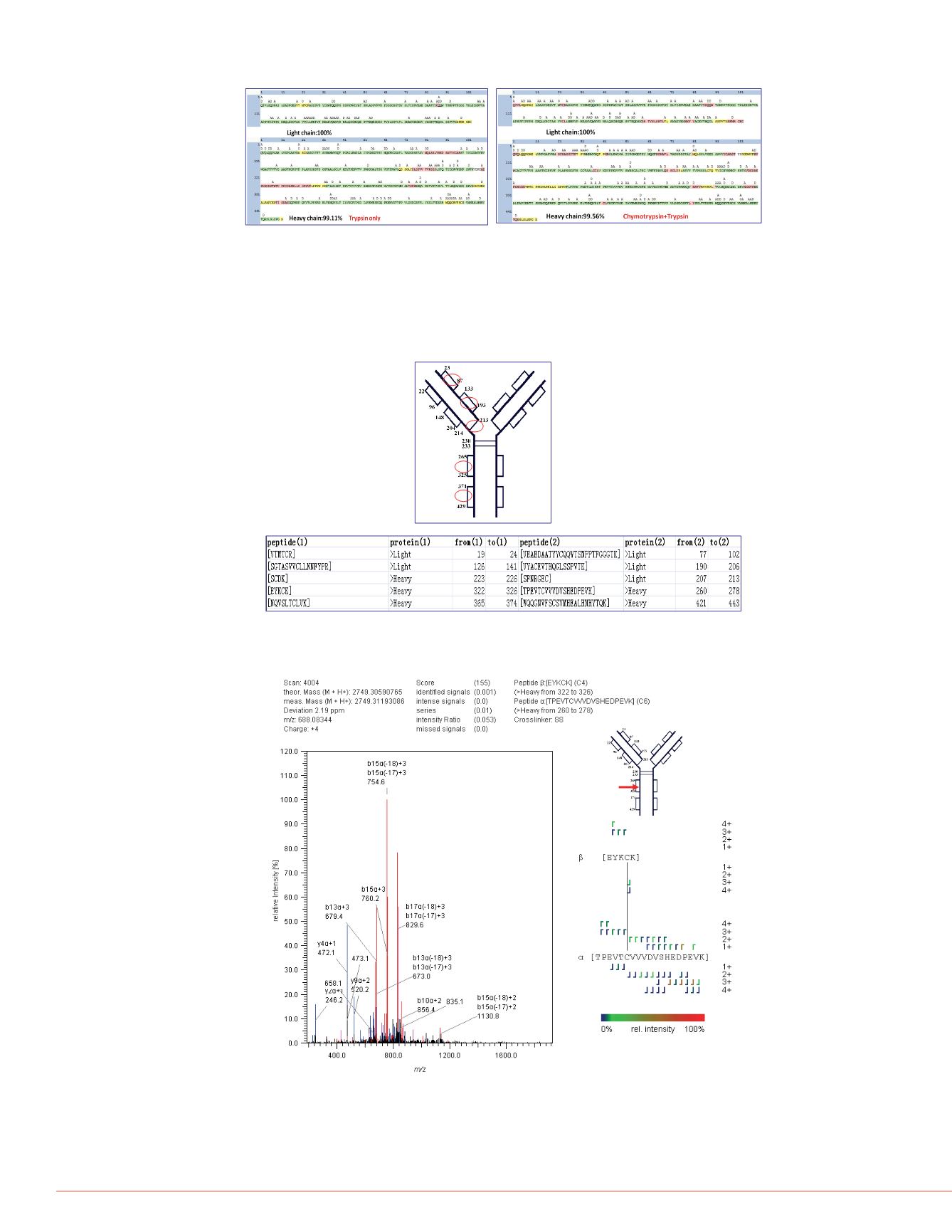
FIGURE 5. The annotated MS/MS spectrum of disulfide bond C265-C325 on
heavy chain (trypsin digested only).
FIGURE 5 is an annotated MS/
Cys325 on heavy chain. The a
indicated that an Orbitrap MS
find that many continuous, disu
These ions are strong evidence
The result of double digested
In our experiments, we also trie
included peptides which were s
and trypsin were used for doubl
coverage of the sample (FIGU
By using the software StavroX
T
digested Rituximab. FIGURE 6
double digested sample.
FIGURE 6. All of the identifie


