Source CID
The source CID (SID) parameter is a DC offset (0–100 eV)
that is added to the source DC offset. The source DC offset
consists of three voltages: capillary DC, S-lens DC, and S-lens
exit lens. The application of this DC offset by setting the
source CID parameter results in collisions of the analytes
inside the injection flatapole with residual gas molecules
present in the source region of the instrument.
All-Ion Fragmentation
All-ion fragmentation (AIF) is a fragmentation type in which
all ions generated in the source are guided through the ion
optics of the mass spectrometer, accumulated in the C-trap,
and sent together to the higher-energy collisional dissociation
(HCD) cell for fragmentation. In this case, the quadrupole is
not set to select a particular precursor but operated in
RF-only pass-through mode. For the analysis of intact
proteins, this is a useful method since different charge states
often show different fragmentation behavior and it is not easy
to predict which one works best.
Data Analysis
Full MS spectra were deconvoluted using Thermo Scientific
™
Protein Deconvolution
™
software version 2.0. From the intact
antibody and the intact heavy chain, the spectra acquired at a
resolution setting of 17,500 were deconvoluted using the
ReSpect
™
algorithm. High resolution spectra from the intact
light chain acquired at a resolution of 140,000 and top-down
spectra acquired at 70,000 resolution were deconvoluted
using the Xtract algorithm. To identify glycoforms of the
intact antibody and the intact heavy chain obtained after
reduction, the masses were compared to the expected masses
with the various combinations of commonly found
glycoforms.
The top-down HCD and AIF spectra were deconvoluted
using the Xtract algorithm in the Thermo Scientific
™
Qual Browser
™
utility. Fragment ion assignment was
performed using Thermo Scientific
™
ProSightPC
™
software
version 3.0 in single protein mode with a fragment ion
tolerance of 5 ppm.
The dataset obtained from the proteolytic digest was
processed with Thermo Scientific
™
Proteome Discoverer
™
software version 1.4, using the SEQUEST
®
algorithm.
A three-protein-entry database was used consisting of the
light chain, the heavy chain in two variants carrying either
Ala or Val at position 219, and trypsin. Mass tolerances were
set to 10 ppm for the precursor and 20 mmu for the fragment
ions. Four variable modifications were considered: carbamid-
omethylation (Cys), oxidation (Met), deamidation (N, Q),
Gln to pyro-Glu conversion, and N,N-dimethylation (Lys)
(relevant for identification of trypsin autolysis products only).
Results and Discussion
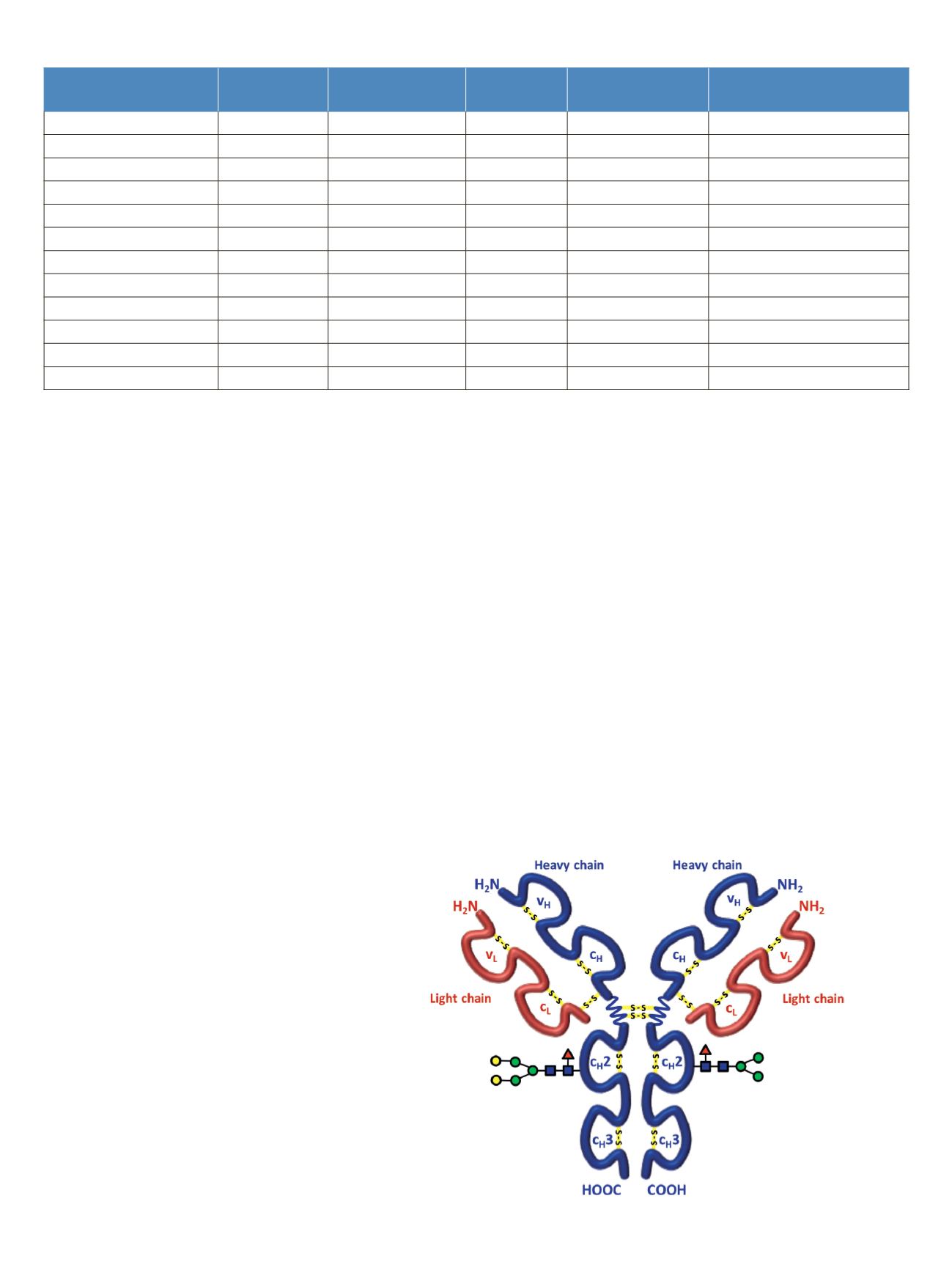
Rituximab is an IgG1 class chimeric monoclonal antibody
against the protein CD20, which consists of two light chains
with 213 amino acids and two heavy chains with 451 amino
acids each in length. The light and heavy chains are connected
via 12 intrachain and 4 interchain disulfide linkages
(Figure 1). The antibody is decorated with glycan structures
attached to residue Asn
301
of each of the two heavy chains.
The composition and length of the attached glycans is quite
diverse, resulting in a microheterogeneity of the molecule.
The variety and relative abundance of the different
glycostructures is essential for the efficiency of the antibody
as a biological drug. The nomenclature of common glycans
attached to antibodies are listed in Figure 2.
3
Table 3. Mass spectrometric parameters used for all experiments
Figure 1. Schematic of molecular structure for the humanized IgG1 class
monoclonal antibody rituximab
Intact Antibody Reduced Antibody Top Down AIF
5-plex MS/MS
(Targeted MS
2
)
Antibody Digest
Method type
Full MS
Full MS (2 segments)
Full MS-AIF
Targeted MS
2
Full MS-dd top 10 HCD
Total run time
30 min
0–15.8/15.8–30 min
25 min
25 min
40 min
Scan range
m/z
1800–5000 800–3500/700–2500 300–2500
Fixed first mass 300
350–2000
Resolution (full MS/MS
2
)
17,500/x
140,000/17,500
70,000
n.a./70,000
70,000/17,500
AGC Full MS
3.00 x 10
6
3.00 x 10
6
3.00 x 10
6
5.00 x 10
5
3.00 x 10
6
(MS)/1.00 x 10
5
(MS
2
)
Max inject time (Full MS/MS
2
)
150 ms
150 ms/200 ms
150 ms
150 ms
100 ms/100 ms
Isolation window
n.a.
n.a.
n.a.
10 Th
2 Th
Microscans
10
5
5
5
1
Capillary temperature
275 °C
275 °C
275 °C
275 °C
275 °C
S-lens RF level
80
80
50
50
50
SID [eV]
80
0/60
n.a.
LC 0/HC 20
n.a.
NCE [%]
n.a.
n.a.
10 to 30
10 to 30
25


