
4
Improving Throughput for Highly Multiplexed Targeted Quanti cation Methods Using Novel API-Remote Instrument Control and State-Model
Data Acquisition Schemes
Results
Highly multiplexed targeted protein quantification requires significant steps of method
refinement prior to implementation. While the determination of proteins is relatively
straightforward based on biology, the selection of peptides as surrogate biomarkers
and corresponding
m/z
values (precursor and product ions) used to uniquely identify
and quantitate the peptide targets becomes challenging. Generally, retention times
and acquisition windows must be determined to maximize instrument cycle time to
achieve robust quantification. To expedite complex experimental method development,
we have created a unique spectral library procedure based on an analytically rigorous
discovery data acquisition scheme. The local spectral library contains both LC and MS
information that can be readily enlisted to build robust methods requiring few
refinement steps.
To first test our methods, a protein mix was spiked in equine plasma (containing PTRC
kit). Spectral library was first built on the neat protein mixture. Experiments performed
on the quadrupole Orbitrap mass spectrometer facilitate unique product ion collection
and detection schemes to not only increase data acquisition, but perform state-model
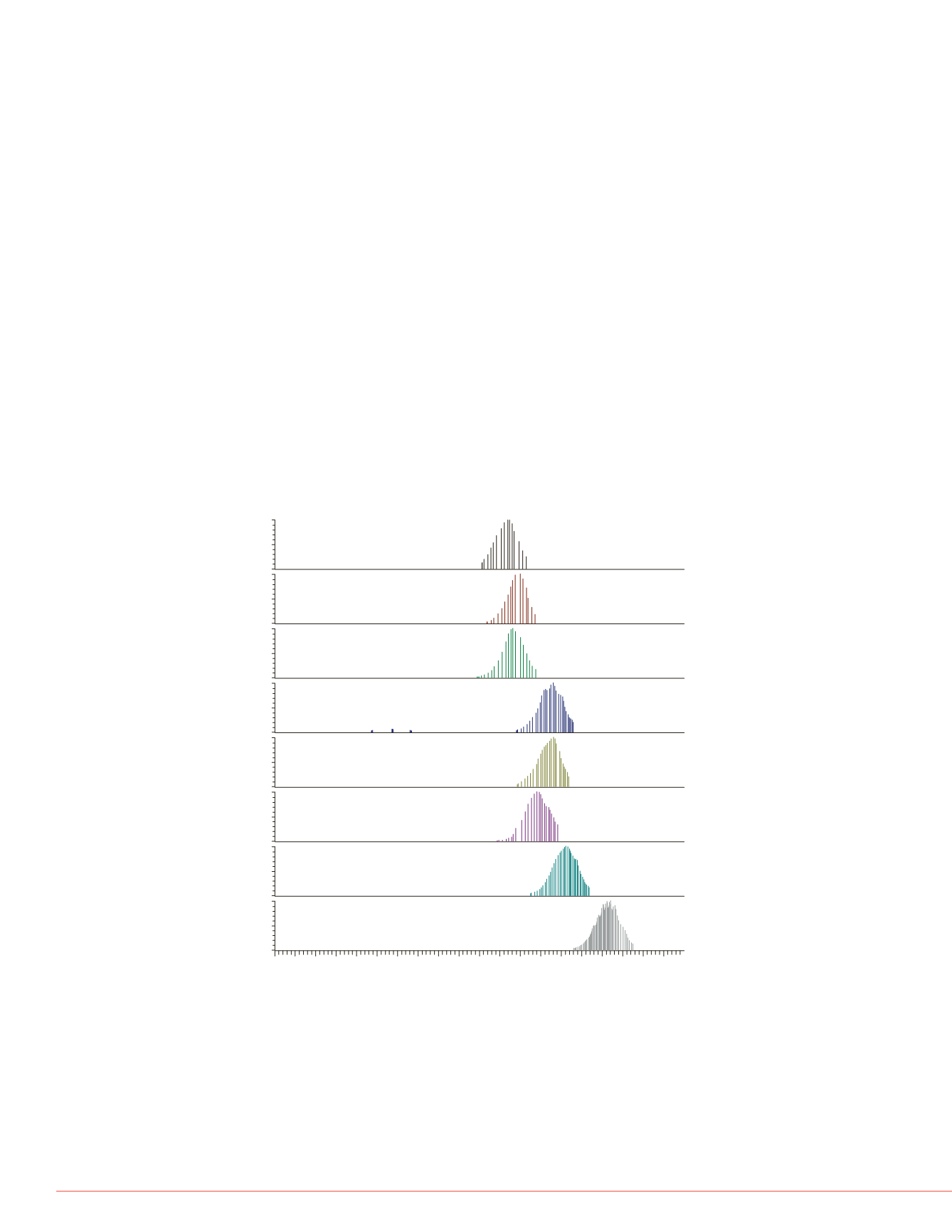
data acquisition, increasing the ability for quantification. Figure 3 shows the result of
the data acquisition scheme, with MS/MS events acquired for the various peptides,
showing the benefit of increased efficiency of triggering events. Figure 4 shows the
distribution of the retention of the various peptides, and as expected, most peptides
elute in the middle of the gradient. Figure 5 shows the CV distribution for the peptides
over four acquisitions (by summing the area of top eight product ions).
ata acquisition schemes for
d scanning window, target
n spectral acquisition. Both
performed to increase the
antification based on high IQ
isotope
nse isotope
e (min)
Stop time for “watch list”
Spectral
Library
Experimental
Spectrum
0
5
10
15
20
25
30
Frequency
SDLYVSDAFHK
2.12E5
SGSAC*VDTPEEGYHAVAVVK
9.24E5
HSSFVNVHLPK
4.06E5
DGGIDPLVR
4.77E5
SSGSLLNNAIK
4.98E5
DVLMSIR
8.33E6
QTVSWAVTPK
1.75E5
EPQVYLAPHR
5.03E5
26.0 26.1 26.2 26.3 26.4 26.5 26.6 26.7 26.8 26.9 27.0 27.1 27.2 27.3 27.4 27.5 27.6 27.7 27.8 27.9
Time (min)
0
50
100
0
50
100
0
50
100
0
50
100
0
50
100
0
50
100
0
50
100
0
50
100
FIGURE 3. Result from high IQ data acquisition scheme for a small list of
peptides. The graphs show the MS/MS events for the various peptides, and
the effective gain in duty cycle.
FIGURE 5. CV distribution fo
FIGURE 4. The number of tar
K562 Cell Line
2,100 proteins were selected fr
algorithm. The algorithm utilize
peptides and create precursor
qualitative and quantitative an
range digest was created.
Figure 6 shows an example wh
full scan MS1 (panel A), but co
zoom-in, panel C).
Determine targeted protein list:
Discovery experiments
Pathway determination
Functional groups
ted acquisition methods from spectral
typic peptides
al precursor and product ion
m/z
values
tribution/relative abundance
tion time windows
relative/absolute quantification across
echnical or biological replicates



















