
6
High-Resolution, Accurate-Mass (HR/AM) and Intelligent Acquisition-Enabled Global Discovery and Quanti cation of Histones, Histone PTMS, and Histone
Modi cation Enzymes in Mesenchymal Stem Cells
Conclusion
We have developed a real-time, intelligent acquisition strategy for HR/AM global
targeted quantification of histones, histone PTMs, and histone modification
enzymes in primary human stem cells upon acute DNA damage and during drug-
evoked senescence.
Assessment of the histone H1, H2a, H2B, H4, and H3 family member abundance
in the soluble nuclear fraction of the cells subjected to genotoxic drug-induced
senescence (5 days after exposure to bleomycin), demonstrated that the
dynamics of histone binding in the senescent cells changes significantly (Figure
3B). Our data also suggest that the composition of the nucleosomal particles
undergoes dramatic changes upon senescence. We have observed significant
reduction in chromatin-bound histone H4 and all of the members of the H1 family
(Figure 3B). These data suggest that with reduction of H1 histone family members
in the chromatin of senescent cells, the overall compaction of chromatin fiber
decreases dramatically.
The representation of macro H2A histone does not change with senescent-
specific transformation. Levels of H2B type1-A and type1-B histones also do not
change significantly. These data suggest histone type specificity in chromatin
dynamics upon reaching a senescent state.
A dramatic increase in chromatin-bound HAT p300 and HMT MLL3 correlates
well with the loss of histone 1 family members from the chromatin. These data
support the hypothesis that relaxation of chromatin compaction in senescent cells
might be a leading cause of increased transcriptional noise (transcriptional
leakage) upon senescence.
References
1. Lunyak, V.V.; Tollervey, J.R. Epigenetics: judge, jury and executioner of stem cell fate.
Epigenetics
. 2012 Aug;7(8):823–40. doi: 10.4161/epi.21141. Epub 2012 Jul 18.
2. Lopez, M.F.; Tollervey, J.; Krastins, B.; Garces, A.; Sarracino, D.; Prakash, A.; Vogelsang,
M.; Geesman, G.; Valderrama, A.; Jordan, I.K.; Lunyak, V.V. Depletion of nuclear histone
H2A variants is associated with chronic DNA damage signaling upon drug-evoked
senescence of human somatic cells. Aging (Albany NY). 2012 Nov;4(11):823–42.
ciated histones and histone
th acute DNA damage (2 hr
cellular senescence
atment).
histones and histone
th acute DNA damage
duced cellular senescence
atment).
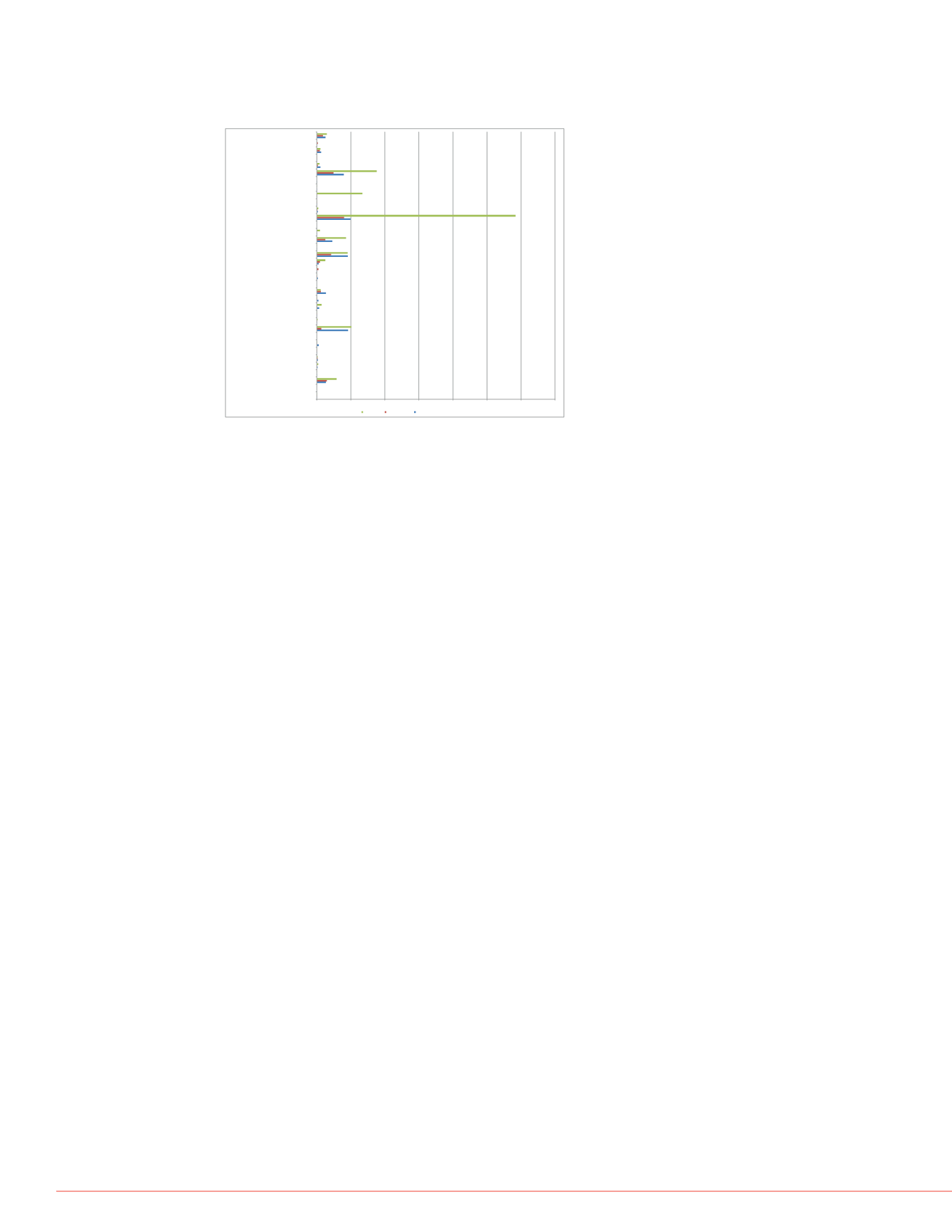
FIGURE 5 Differential abundance of cytoplasmic fraction histones and histone
modification proteins in mesenchymal stem cells with acute DNA damage
(2 hr treatment with bleomycin) and DNA damage-induced cellular senescence
(cellular recovery after 5 days of post-bleomycin treatment).
0.00E+00
1.00E+08
2.00E+08
3.00E+08
raseMLL3 (Fragment)
Histone deacetylase 5
Histone deacetylase 4
ethyltransferaseMLL2
deacetylase inducible)
ome protein2-like 1 (S.
acetyltransferase p300
-binding proteinRBBP4
Histone deacetylase 1
nsferase H3 lysine-79
Histone deacetylase 2
thyltransferase PRDM7
Area
HistoneModificationProteins
Nuclear Fraction
Bleomycin+5Days
vs
NoTreatment
NoTreatment
Bleomycin+ 5Days
0.00E+00
2.00E+08
4.00E+08
6.00E+08
H1.0
H3.1
1-B
1-A
/G/I
2-B
H1.3
H1.2
H1x
eH4
2-C
1-D
-B/E
H1.5
1-A
2A.V
2A.1
2A.2
3.1t
H1.4
1-H
H1.1
ne2
Area
HistonesNuclear Fraction
Bleomycin+5days
vs
No treatment
clearNo Treatment
NuclearBleomycin= 5Days
0.00E+00
1.00E+08
2.00E+08
3.00E+08
4.00E+08
5.00E+08
6.00E+08
7.00E+08
HUMANHistoneH1.0
HUMANHistoneH3.1
HUMANHistoneH2B type 1-B
HUMANHistoneH2B type 1-A
HUMANHistoneH2B type 1-C/E/F/G/I
HUMANHistoneH2A type 2-B
HUMANHistoneH1.3
HUMANHistoneH1.2
HUMANHistoneH1x
HUMANHistoneH4
HUMAN Putative histoneH2B type 2-C
HUMANHistoneH2A type 1-D
HUMANHistoneH2A type 1-B/E
HUMANHistoneH1.5
HUMANHistoneH2A type 1-A
HUMANHistoneH2A.V
HUMAN Corehistonemacro-H2A.1
HUMAN Corehistonemacro-H2A.2
HUMANHistoneH3.1t
HUMANHistoneH1.4
HUMANHistoneH2B type 1-H
HUMANHistoneH1.1
HUMANHistone cluster 2 H3 pseudogene2
HUMANHistone-lysineN-methyltransferaseMLL3 (Fragment)
HUMANHistone deacetylase 5
HUMANHistone deacetylase 4
HUMANHistone-lysineN-methyltransferaseMLL2
HUMAN Keratin23 (Histone deacetylase inducible)
HUMANNHP2non-histone chromosome protein2-like 1 (S.cerevisiae)
HUMANHistone acetyltransferase p300
HUMANHistone-binding proteinRBBP4
HUMANHistone deacetylase 1
HUMANHistone-lysineN-methyltransferase H3 lysine-79 specific
HUMANHistone deacetylase 2
HUMAN Probablehistone-lysine N-methyltransferase PRDM7
Cytoplasmic Fraction
HisonesandHistone ModificationProteins
NoTreatment
Bleomycin+ 5Days
Bleomycin
All trademarks are the property of Thermo Fisher Scientific and its subsidiaries.
This information is not intended to encourage use of these products in any manners that might infringe the
intellectual property rights of others.
00E+00
1.00E+09
2.00E+09
3.00E+09
4.00E+09
Area
HistonesChromatinFraction
Bleomycin+5days
vs
No treatment
NoTreatment
Bleomycin+ 5Days
0.00E+00 2.00E+07 4.00E+07 6.00E+07 8.00E+07 1.00E+08 1.20E+08
feraseMLL3 (Fragment)
NHistone deacetylase 5
NHistone deacetylase 4
methyltransferaseMLL2
e deacetylase inducible)
some protein2-like 1 (S.
acetyltransferase p300
e-binding proteinRBBP4
NHistone deacetylase 1
se H3 lysine-79 specific
NHistone deacetylase 2
ethyltransferase PRDM7
Area
HistoneModificationProteins
ChromatinFraction
Bleomycin+5days
vs
No treatment
NoTreatment
Bleomycin+ 5days



















