
5
Thermo Scienti c Poster Note
•
PN ASMS13_T351_MLopez_E 07/13S
Conclusion
We have developed a rea
targeted quantification of
enzymes in primary huma
evoked senescence.
Assessment of the histon
in the soluble nuclear frac
senescence (5 days after
dynamics of histone bindi
3B). Our data also sugge
undergoes dramatic chan
reduction in chromatin-bo
(Figure 3B). These data s
in the chromatin of senes
decreases dramatically.
The representation of ma
specific transformation. L
change significantly. Thes
dynamics upon reaching
A dramatic increase in ch
well with the loss of histo
support the hypothesis th
might be a leading cause
leakage) upon senescenc
References
1. Lunyak, V.V.; Tollervey, J.R.
Epigenetics
. 2012 Aug;7(8):
2. Lopez, M.F.; Tollervey, J.; K
M.; Geesman, G.; Valderra
H2A variants is associated
senescence of human som
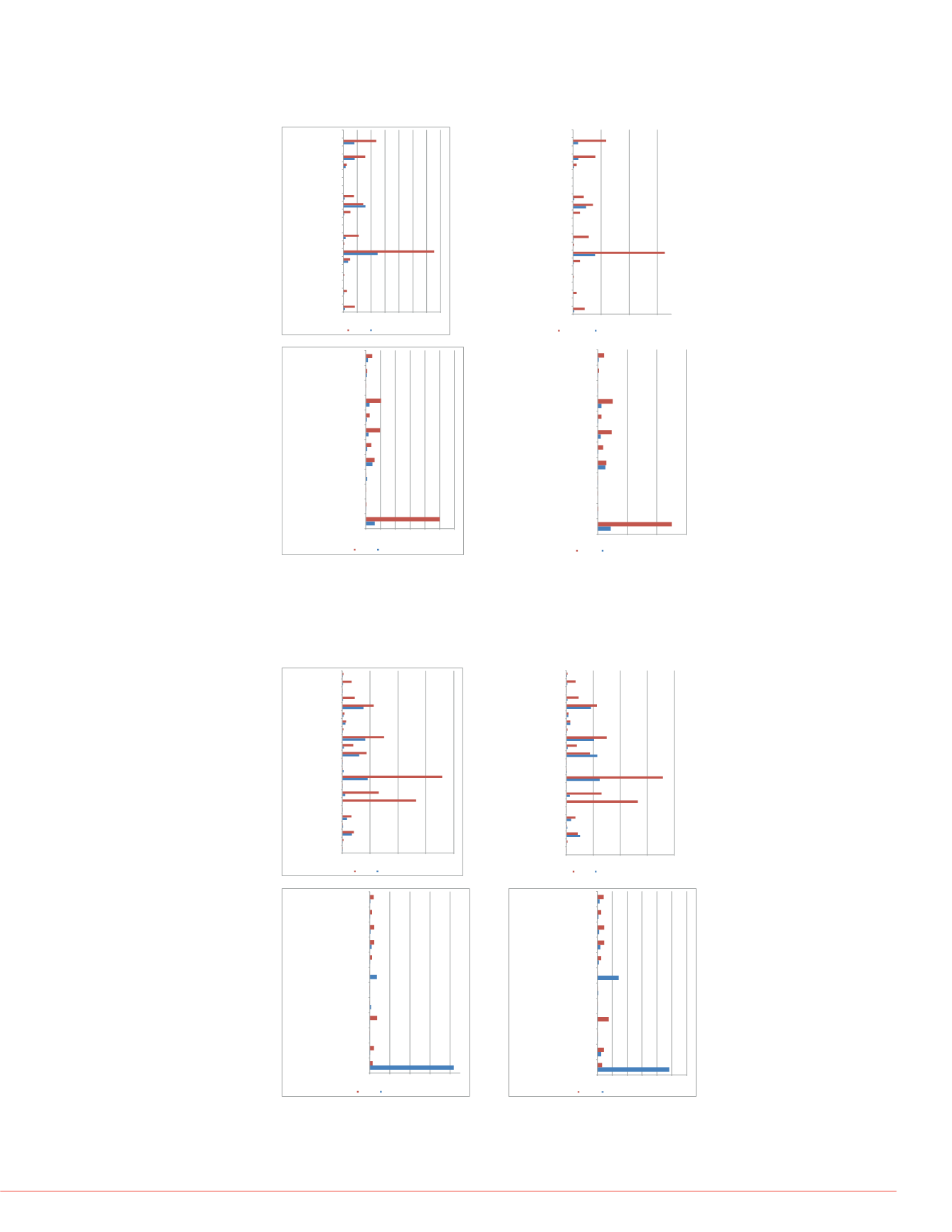
FIGURE 4. Differential abundance of chromatin associated histones and histone
modification proteins in mesenchymal stem cells with acute DNA damage (2 hr
treatment with bleomycin) and DNA damage-induced cellular senescence
(cellular recovery after 5 days of post-bleomycin treatment).
FIGURE 3. Differential abundance of nuclear fraction histones and histone
modification proteins in mesenchymal stem cells with acute DNA damage
(2 hr treatment with bleomycin) and DNA damage-induced cellular senescence
(cellular recovery after 5 days of post-bleomycin treatment).
FIGURE 5 Differential abund
modification proteins in me
(2 hr treatment with bleomy
(cellular recovery after 5 da
0.00E+00 1.00E+08 2.00E+08 3.00E+08 4.00E+08 5.00E+08 6.00E+08 7.00E+08
HUMANHistoneH1.0
HUMANHistoneH3.1
HUMANHistoneH2B type 1-B
HUMANHistoneH2B type 1-A
HUMANHistoneH2B type 1-C/E/F/G/I
HUMANHistoneH2A type 2-B
HUMANHistoneH1.3
HUMANHistoneH1.2
HUMANHistoneH1x
HUMANHistoneH4
HUMAN Putative histoneH2B type 2-C
HUMANHistoneH2A type 1-D
HUMANHistoneH2A type 1-B/E
HUMANHistoneH1.5
HUMANHistoneH2A type 1-A
HUMANHistoneH2A.V
HUMAN Corehistonemacro-H2A.1
HUMAN Corehistonemacro-H2A.2
HUMANHistoneH3.1t
HUMANHistoneH1.4
HUMANHistoneH2B type 1-H
HUMANHistoneH1.1
HUMANHistone cluster 2 H3 pseudogene2
AxisTitle
HistonesNuclearFraction
Bleomycin
vs
No treatment
NoTreatment
Bleomycin
0.00E+00 5.00E+07 1.00E+08 1.50E+08 2.00E+08 2.50E+08 3.00E+08
HUMANHistone-lysineN-methyltransferaseMLL3 (Fragment)
HUMANHistone deacetylase 5
HUMANHistone deacetylase 4
HUMANHistone-lysineN-methyltransferaseMLL2
HUMAN Keratin23 (Histone deacetylase inducible)
HUMANNHP2non-histone chromosome protein2-like 1 (S.
cerevisiae)
HUMANHistone acetyltransferase p300
HUMANHistone-binding proteinRBBP4
HUMANHistone deacetylase 1
HUMANHistone-lysineN-methyltransferase H3 lysine-79
specific
HUMANHistone deacetylase 2
HUMAN Probablehistone-lysine N-methyltransferase PRDM7
Area
HistoneModificationProteinsNuclear
Fraction
Bleomycin
vs
NoTreatment
NoTreatment
Bleomycin
0.00E+00
1.00E+08
2.00E+08
3.00E+08
HUMANHistone-lysineN-methyltransferaseMLL3 (Fragment)
HUMANHistone deacetylase 5
HUMANHistone deacetylase 4
HUMANHistone-lysineN-methyltransferaseMLL2
HUMAN Keratin23 (Histone deacetylase inducible)
HUMANNHP2non-histone chromosome protein2-like 1 (S.
cerevisiae)
HUMANHistone acetyltransferase p300
HUMANHistone-binding proteinRBBP4
HUMANHistone deacetylase 1
HUMANHistone-lysineN-methyltransferase H3 lysine-79
specific
HUMANHistone deacetylase 2
HUMAN Probablehistone-lysine N-methyltransferase PRDM7
Area
HistoneModificationProteins
Nuclear Fraction
Bleomycin+5Days
vs
NoTreatment
NoTreatment
Bleomycin+ 5Days
0.00E+00
2.00E+08
4.00E+08
6.00E+08
HUMANHistoneH1.0
HUMANHistoneH3.1
HUMANHistoneH2B type 1-B
HUMANHistoneH2B type 1-A
HUMANHistoneH2B type 1-C/E/F/G/I
HUMANHistoneH2A type 2-B
HUMANHistoneH1.3
HUMANHistoneH1.2
HUMANHistoneH1x
HUMANHistoneH4
HUMAN Putative histoneH2B type 2-C
HUMANHistoneH2A type 1-D
HUMANHistoneH2A type 1-B/E
HUMANHistoneH1.5
HUMANHistoneH2A type 1-A
HUMANHistoneH2A.V
HUMAN Corehistonemacro-H2A.1
HUMAN Corehistonemacro-H2A.2
HUMANHistoneH3.1t
HUMANHistoneH1.4
HUMANHistoneH2B type 1-H
HUMANHistoneH1.1
HUMANHistone cluster 2 H3 pseudogene2
Area
HistonesNuclear Fraction
Bleomycin+5days
vs
No treatment
NuclearNo Treatment
NuclearBleomycin= 5Days
A
D
C
B
nce
]EVDR
LR
l]FNLTVK
ethyl]GSGILSFFAAQ[Deamid]AGAR
IAS[Phosphoryl]TEVKLR
]AAR
R
R[Methyl]
NDIFER
NDIFER
]NSFVNDIFER
]NSFVNDIFER[Methyl]
TAVRLLLPGELAK
42]FVNDIFER
42]FVNDIFER
NSFVNDIFER
]NSFVNDIFER
42]FVN[Deamid]DIFER
amid]S[S_42]FVNDIFER
42]FVNDIFER[Methyl]
]NS[S_42]FVNDIFER
[Oxid]NSFVNDIFER
[Oxid]NS[S_42]FVNDIFER
FVNDIFER
N[Deamid]DIFER
N[Deamid]DIFER
NDIFER[Methyl]
]N[Deamid]S[S_42]FVNDIFER
[Oxid]N[Deamid]SFVNDIFER
SK
K
eamid]DEELNK
APAEK
APAEK
APPAEK
Phosphoryl]K[di-Methyl]K[Methyl]
[Deamid]HC[Carboxymethyl]ISSDPR
]GVSRPVIAC[Carboxymethyl]SVTIK
_42]R
DIFER
amid]SFLN[Deamid]DIFER
]NSFLNDIFER
R
VYALKR
PAIR
PAIR
hyl]
]DVVYALK
]DVVYALK
cet]R
42]KR
YALK
ethyl]R
ALKR
d]DVVYALK
d]DVVYALK
d]DVVYALK
d]DVVYALK
]DVVYALKR
yl]
yl]
eamid]AVLLPK
mid]IQAVLLPK
LPNIQAVLLPK
mid]IQ[Deamid]AVLLPK
LPNIQ[Deamid]AVLLPK
eamid]AVLLPK[Methyl]
eamid]AVLLPKK
mid]IQAVLLPKK
LPNIQAVLLPKK
mid]IQAVLLPK[Methyl]
thyl]GEGSR
l]
DEELNK
IQ[Deamid]AVLLPK
[Deamid]IQAVLLPK
]GGVLPNIQAVLLPK
]LALADDK
]LALADDKK
HGLPAK
LR
Methyl]TDLR
l]TDLR
AR
PATGGVK
tri-Methyl]K[di-Methyl]
PAPAEK
0.00E+00
1.00E+08
2.00E+0
HUMANHistoneH1.0
HUMANHistoneH3.1
HUMANHistoneH2B type 1-B
HUMANHistoneH2B type 1-A
HUMANHistoneH2B type 1-C/E/F/G/I
HUMANHistoneH2A type 2-B
HUMANHistoneH1.3
HUMANHistoneH1.2
HUMANHistoneH1x
HUMANHistoneH4
HUMAN Putative histoneH2B type 2-C
HUMANHistoneH2A type 1-D
HUMANHistoneH2A type 1-B/E
HUMANHistoneH1.5
HUMANHistoneH2A type 1-A
HUMANHistoneH2A.V
HUMAN Corehistonemacro-H2A.1
HUMAN Corehistonemacro-H2A.2
HUMANHistoneH3.1t
HUMANHistoneH1.4
HUMANHistoneH2B type 1-H
HUMANHistoneH1.1
HUMANHistone cluster 2 H3 pseudogene2
HUMANHistone-lysineN-methyltransferaseMLL3 (Fragment)
HUMANHistone deacetylase 5
HUMANHistone deacetylase 4
HUMANHistone-lysineN-methyltransferaseMLL2
HUMAN Keratin23 (Histone deacetylase inducible)
HUMANNHP2non-histone chromosome protein2-like 1 (S.cerevisiae)
HUMANHistone acetyltransferase p300
HUMANHistone-binding proteinRBBP4
HUMANHistone deacetylase 1
HUMANHistone-lysineN-methyltransferase H3 lysine-79 specific
HUMANHistone deacetylase 2
HUMAN Probablehistone-lysine N-methyltransferase PRDM7
NoTreatment
Bl
All trademarks are the property of Ther
This information is not intended to enco
intellectual property rights of others.
0.00E+00
1.00E+09
2.00E+09
3.00E+09
4.00E+09
HUMANHistoneH1.0
HUMANHistoneH3.1
HUMANHistoneH2B type 1-B
HUMANHistoneH2B type 1-A
HUMANHistoneH2B type 1-C/E/F/G/I
HUMANHistoneH2A type 2-B
HUMANHistoneH1.3
HUMANHistoneH1.2
HUMANHistoneH1x
HUMANHistoneH4
HUMAN Putative histoneH2B type 2-C
HUMANHistoneH2A type 1-D
HUMANHistoneH2A type 1-B/E
HUMANHistoneH1.5
HUMANHistoneH2A type 1-A
HUMANHistoneH2A.V
HUMAN Corehistonemacro-H2A.1
HUMAN Corehistonemacro-H2A.2
HUMANHistoneH3.1t
HUMANHistoneH1.4
HUMANHistoneH2B type 1-H
HUMANHistoneH1.1
HUMANHistone cluster 2 H3 pseudogene2
Area
HistonesChromatinFraction
Bleomycin
vs
NoTreatment
NoTreatment
Bleomycin
0.00E+00
1.00E+09
2.00E+09
3.00E+09
4.00E+09
HUMANHistoneH1.0
HUMANHistoneH3.1
HUMANHistoneH2B type 1-B
HUMANHistoneH2B type 1-A
HUMANHistoneH2B type 1-C/E/F/G/I
HUMANHistoneH2A type 2-B
HUMANHistoneH1.3
HUMANHistoneH1.2
HUMANHistoneH1x
HUMANHistoneH4
HUMAN Putative histoneH2B type 2-C
HUMANHistoneH2A type 1-D
HUMANHistoneH2A type 1-B/E
HUMANHistoneH1.5
HUMANHistoneH2A type 1-A
HUMANHistoneH2A.V
HUMAN Corehistonemacro-H2A.1
HUMAN Corehistonemacro-H2A.2
HUMANHistoneH3.1t
HUMANHistoneH1.4
HUMANHistoneH2B type 1-H
HUMANHistoneH1.1
HUMANHistone cluster 2 H3 pseudogene2
Area
HistonesChromatinFraction
Bleomycin+5days
vs
No treatment
NoTreatment
Bleomycin+ 5Days
0.00E+00
4.00E+07
8.00E+07
1.20E+08
1.60E+08
HUMANHistone-lysineN-methyltransferaseMLL3 (Fragment)
HUMANHistone deacetylase 5
HUMANHistone deacetylase 4
HUMANHistone-lysineN-methyltransferaseMLL2
HUMAN Keratin23 (Histone deacetylase inducible)
HUMANNHP2non-histone chromosome protein2-like 1 (S.
cerevisiae)
HUMANHistone acetyltransferase p300
HUMANHistone-binding proteinRBBP4
HUMANHistone deacetylase 1
HUMANHistone-lysineN-methyltransferase H3 lysine-79 specific
HUMANHistone deacetylase 2
HUMAN Probablehistone-lysine N-methyltransferase PRDM7
Area
HistoneModificationProteinsChromatin
Fraction
Bleomycin
vs
NoTreatment
NoTreatment
Bleomycin
0.00E+00 2.00E+07 4.00E+07 6.00E+07 8.00E+07 1.00E+08 1.20E+08
HUMANHistone-lysineN-methyltransferaseMLL3 (Fragment)
HUMANHistone deacetylase 5
HUMANHistone deacetylase 4
HUMANHistone-lysineN-methyltransferaseMLL2
HUMAN Keratin23 (Histone deacetylase inducible)
HUMANNHP2non-histone chromosome protein2-like 1 (S.
cerevisiae)
HUMANHistone acetyltransferase p300
HUMANHistone-binding proteinRBBP4
HUMANHistone deacetylase 1
HUMANHistone-lysineN-methyltransferase H3 lysine-79 specific
HUMANHistone deacetylase 2
HUMAN Probablehistone-lysine N-methyltransferase PRDM7
Area
HistoneModificationProteins
ChromatinFraction
Bleomycin+5days
vs
No treatment
NoTreatment
Bleomycin+ 5days
A
D
C
B



















