
4
Comprehensive Peptide Searching Work ow to Maximize Protein Identi cations
Search).
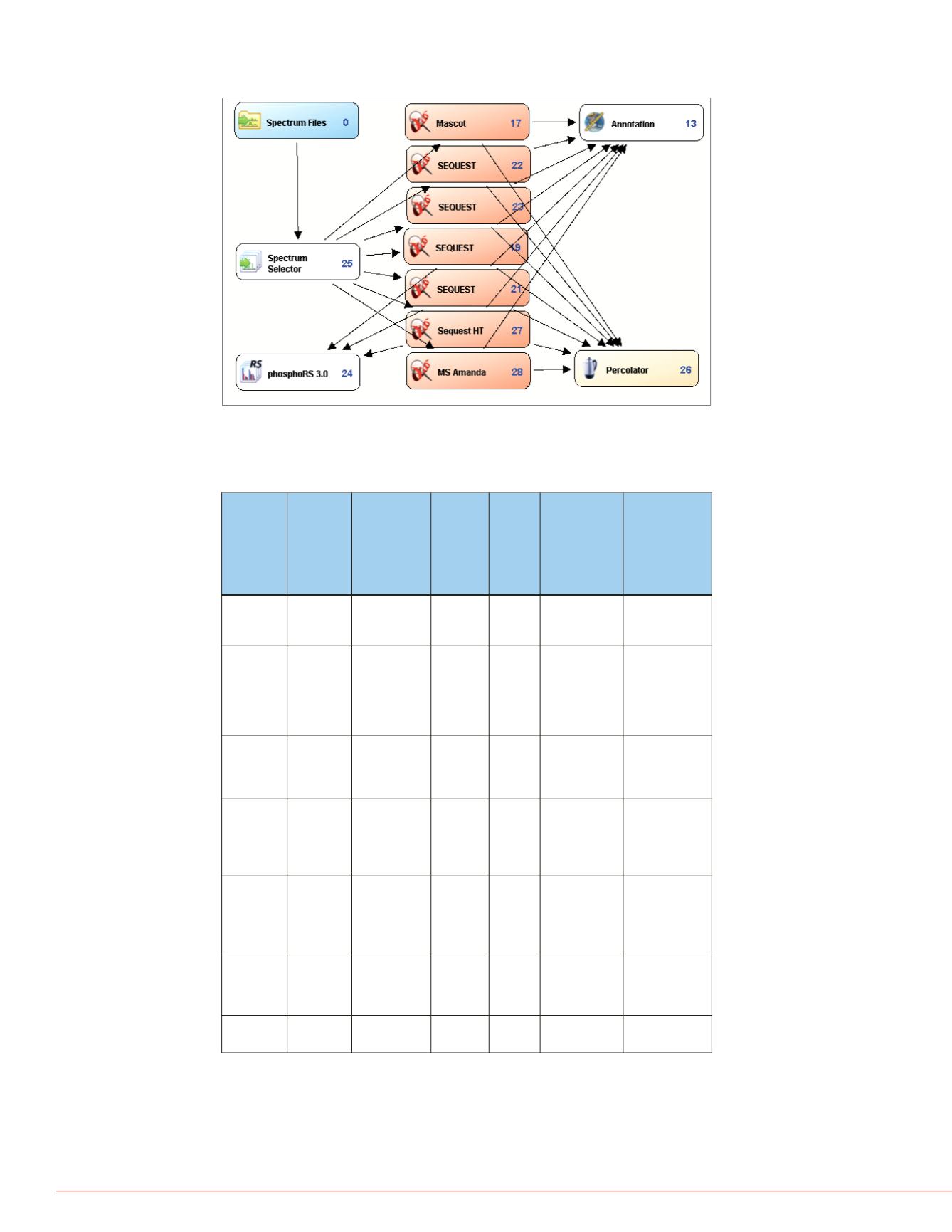
FIGURE 1. Structure of the comprehensive workflow
eptide Searching Workflow to Maximize Protein
1
, David Sarracino
1
, Bryan Krastins
1
, MingMing Ning
2
, Barbara Frewen
1
, Scott Peterm
adali
1
, Jennifer Sutton
1
, Mary F. Lopez
1
S (Biomarker Research in Mass Spectrometry), Cambridge, MA;
2
Massachusetts Gen
rotein
ence
anslational
database search
rch engines
S Amanda)
chosen for each
nces from high
1
.
h-confidence,
fications
orkflow
.
ass
od in biological
in identifications
any factors,
S data
ion of spectra
ptide matches
everal factors
conventional
ate combination
nd iterative
identify
ensive MS/MS
teome
ence
rious search
explored. We
of protein
igestion and
TABLE 1. Parameters and modifications used in
comprehensive search workflow
Search
Engine
Precursor
Mass
Tolerance
Fragment
Mass
Tolerance
(Q Exactive
MS/LTQ
Orbitrap
Velos MS)
Missed
Cleavage
Enzyme
Static
Modification
Dynamic
Modification
Mascot
5 ppm
0.02 Da /
0.4 Da
2
Semi
Trypsin
Carboxymethyl
(C)
Oxidation (M);
Acetyl (K);
Methyl (K)
SEQUEST
5 ppm
0.02 Da /
0.4 Da
3
Trypsin
(Full)
Carboxymethyl
(C)
Oxidation (M);
ADP-Ribosyl
(N,R); Myristoyl
(K); Deamidation
(N,Q); Phospho
(S)
SEQUEST
5 ppm
0.02 Da /
0.4 Da
3
Trypsin
(Full)
Carboxymethyl
(C)
Oxidation (M);
Dioxidation (M);
Trimethyl (K,R);
Phospho (S,T)
SEQUEST
5 ppm
0.02 Da /
0.4 Da
3
Trypsin
(Full)
Carboxymethyl
(C)
Oxidation (M);
Carbamyl (K,R);
Deamidated
(N,Q); Amidation
(Any C-Terminus)
SEQUEST
5 ppm
0.02 Da /
0.4 Da
3
Trypsin
(Full)
Carboxymethyl
(C)
Oxidation (M);
Methyl (K,R);
Dimethyl (K,R);
Trimethyl (K,R);
Acetyl (K)
Sequest
HT
5 ppm
0.02 Da /
0.4 Da
3
Trypsin
(Full)
Carboxymethyl
(C)
Oxidation (M);
Phospho (S,T,Y);
Deamidated
(N,Q);
MS
Amenda
5 ppm
0.02 Da /
0.4 Da
3
Trypsin
(Full)
Carboxymethyl
(C)
Oxidation (M);
Acetyl (K)
Results
We compared the results from our comprehensive searching
strategy with a standard search strategy. We found that on
average, the nu ber of high-confidence peptide identifications
(FDR≤0.01) increased approximately 2
-fold with our
We further investigat
spectra in the data se
comprehensive searc
of matched spectra i
comprehensive searc
Sequence
CCKHPEAKRMPCAEDYLSVVL
VLHEK
CYAKVFDEFKPLVEEPQNLIK
DKDEAEQAVSR
LVRPEVDVMCTAFHDNEETFLK
INNEDNSQFK
RMPCAEDYLSVVLNQLCVLHE
SEPKWEVVEPLK
TCVADESAENCDK
YYFNCNNWLSKVEGDRQWCR
TABLE 2. Examples
from the comprehe
Table 3. Comparativ
File
Total
Spectra
Matc
Spe
Stan
Sear
(FDR≤
Sample1
27215
28.
Sample2
14005
15.
Sample3
43036
5.1
Sample4
44450
9.5



















