
6
Comprehensive Peptide Searching Work ow to Maximize Protein Identi cations
gmentation.
e Discoverer 1.4
abase using the
d compared with
tions (oxidation
ylation as static
(Standard
workflow
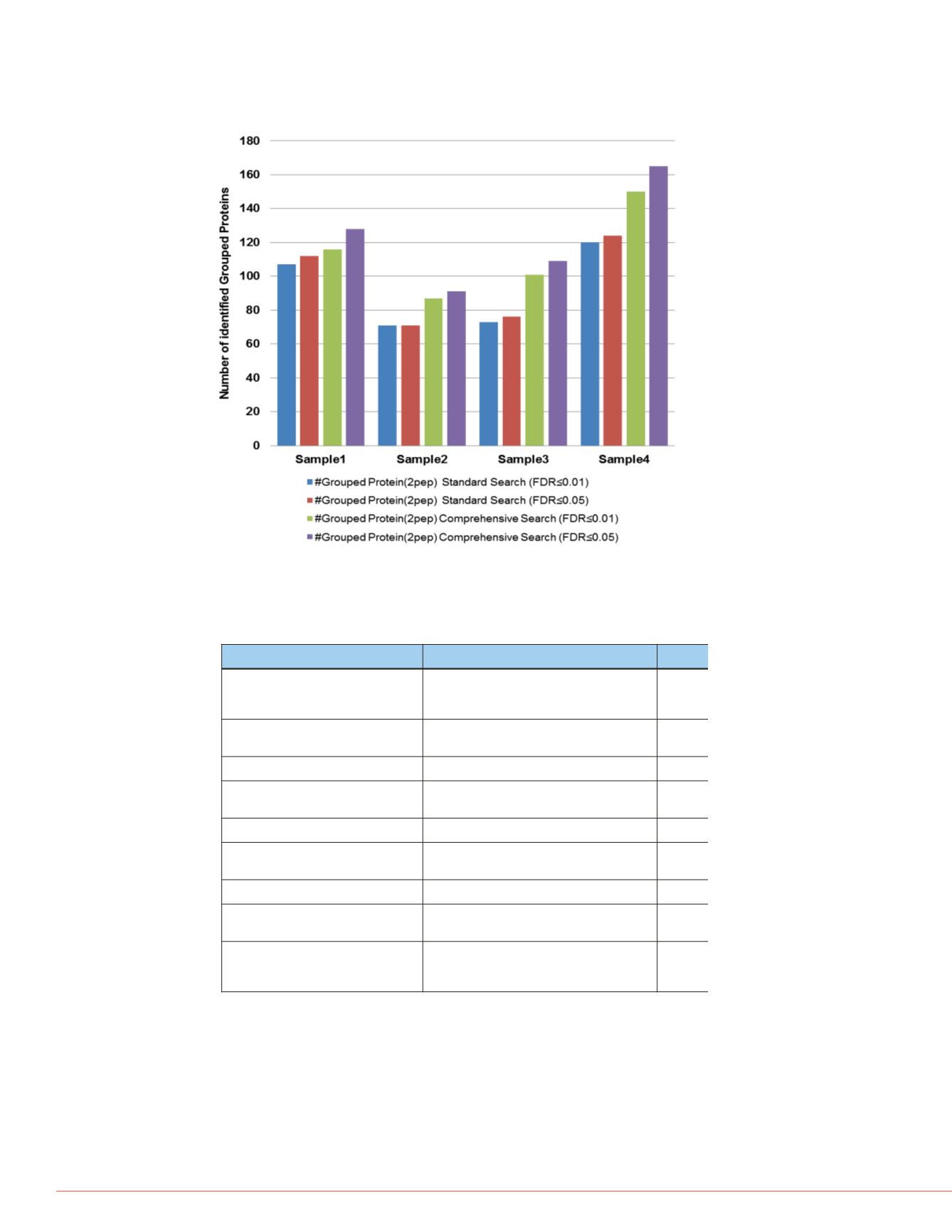
FIGURE 3. The comprehensive workflow increases the
number of identified protein groups with at least two
peptide hits per protein.
when using the co
Proteome Discov
References
1. Khoury GA, Balib
translational modi
curation of the sw
13;1.
2. Schandorff S, Ols
Y, Andersen JS,
database for cSN
4(6):465-6.
SEQUEST, Sequest HT and Percolator
registered trademark of Matrix Science
Group at Research Institute of Molecula
Hagenberg. All other trademarks are the
This information is not intended to enco
property rights of others.
o Maximize Protein Identifications
2
, Barbara Frewen
1
, Scott Peterman
1
, Gregory Byram
1
,
bridge, MA;
2
Massachusetts General Hospital, Boston, MA
ed in
Static
ification
Dynamic
Modification
oxymethyl
(C)
Oxidation (M);
Acetyl (K);
Methyl (K)
oxymethyl
(C)
Oxidation (M);
ADP-Ribosyl
(N,R); Myristoyl
(K); Deamidation
(N,Q); Phospho
(S)
oxymethyl
(C)
Oxidation (M);
Dioxidation (M);
Trimethyl (K,R);
Phospho (S,T)
oxymethyl
(C)
Oxidation (M);
Carbamyl (K,R);
Deamidated
(N,Q); Amidation
(Any C-Terminus)
oxymethyl
(C)
Oxidation (M);
Methyl (K,R);
Dimethyl (K,R);
Trimethyl (K,R);
Acetyl (K)
oxymethyl
(C)
Oxidation (M);
Phospho (S,T,Y);
Deamidated
(N,Q);
oxymethyl
(C)
Oxidation (M);
Acetyl (K)
We further investigate the number of matched and unmatched
spectra in the data sets comparing the standard search and our
comprehensive search strategy. We found that the percentage
of matched spectra improves significantly when using the
comprehensive search workflow (Table 3).
Sequence
Modification
q-Value
CCKHPEAKRMPCAEDYLSVVLNQLC
VLHEK
C1(Carboxymethyl); C2(Carboxymethyl);
K3(Myristoyl); M10(Oxidation);
C12(Carboxymethyl); C25(Carboxymethyl)
≤0.001
CYAKVFDEFKPLVEEPQNLIK
C1(Carboxymethyl); K4(Methyl); K10(Acetyl);
K21(Methyl)
≤0.001
DKDEAEQAVSR
K2(Acetyl); R11(Trimethyl)
0.008
LVRPEVDVMCTAFHDNEETFLKK
R3(Dimethyl); M9(Oxidation);
C10(Carboxymethyl); K22(Acetyl); K23(Acetyl)
0.004
INNEDNSQFK
N3(ADP-Ribosyl); K10(Myristoyl)
0.01
RMPCAEDYLSVVLNQLCVLHEK
R1(Trimethyl); M2(Dioxidation);
C4(Carboxymethyl); C17(Carboxymethyl)
≤0.001
SEPKWEVVEPLK
K4(Trimethyl); K12(Dimethyl)
0.004
TCVADESAENCDK
C2(Carboxymethyl); C11(Carboxymethyl);
K13(Dimethyl)
≤0.001
YYFNCNNWLSKVEGDRQWCR
C5(Carboxymethyl); K11(Methyl);
R16(Trimethyl); C19(Carboxymethyl);
R20(Methyl)
0.006
TABLE 2. Examples of peptides containing multiple PTMs
from the comprehensive search strategy
Table 3. Comparative table for matched spectra
Matched
Matched
Matched



















