
Figure 2 shows the summed SRM chromatograms
for the four targeted rhEPO peptides and the labeled
analogues. The labeled peptide can be used to confirm the
correct elution time as well as the ion ratio provided more
than one transition was used to monitor each peptide.
A level of 500 amol on column was used to test the
detection capabilities of the approach used, which would
equate to a concentration of ca. 1.7 ng/mL. Note that the
responses of T
4
, T
11
, T
17
markers were greater than 10000
counts, indicating lower levels of detection to be about
10x lower (or 0.2 ng/mL) without requiring nanoliter flow
rates, which simplifies the experiment and increases the
robustness of the method.
In addition to establishing the correct retention times
for targeted peptides, the stable-isotope labeled peptides
can be used for correct ion ratio determination as an
additional means of verification. Figure 3 shows
comparative full-scan product ion spectra for the (3A)
unlabeled and (3B) labeled T
11
peptide. Note the y-series
detected for each, providing sequencing information and
site determination for the stable isotope labeled residue
such as the a
2
/b
2
fragments as well as the y
6
for the
unlabeled peptide. The two product ions used for
detecting the T
11
peptide were the y
4
and y
5
ions. The
calculated abundance ratios for the unlabeled and labeled
peptides were ca. 25%. The insets to the right of Figure 3
show the measured ion abundance for each SRM transi-
tion at 500 amol level. The calculated ratio is within
experimental error to be used as an additional means
of confirmation for the targeted peptide elution.
Figure 4 shows the quantification curve calculated
for the controlled rhEPO spiking of horse plasma. The
values show excellent agreement between theoretical and
experimentally determined levels based on the integrated
peak area ratios between the unlabeled and labeled
targeted rhEPO peptides. The %CVs for each was less
than 20% at 500 amol level indicating excellent
capabilities to quantify the presence of rhEPO in plasma.
While a positive confirmation would only require one
diagnostic peptide to be present, this method yields four
proteotypic peptides that could be used unequivocally
to increase the confidence in a positive determination.
The second sample set was used to test the entire
workflow. A female horse (500 kg) was administered
rhEPO intravenously using 8000 IU (0.08 mg/kg) for four
days. Following the injection on the fourth day, blood was
withdrawn at 0, 0.5, 1, 2, 3, 4, 6, 8, 10, 24, 48, and 72
hour intervals. Samples for each time point were processed
using the method outlined previously, reducing complexity
of the resulting protein digest mixture. The protocols of
most horse racing commissions require the saliva, urine,
and/or blood sample to be taken from the winning horse
following completion of a race. The 72 hour time window
represents a possible maximum duration between the final
doping and racing while maintaining a pharmacological
effect following administration of rhEPO/DPO. The 8000
IU dose is also an estimate of the dose required to induce
the desired biological effects of increasing oxygen carrying
capacity for equine athletes. The proposed protocol must
enable a reduction of sample loss through the number of
sample purification, filtering, reconstitution, and digestion
steps prior to mass spectral analysis. Figure 5 shows the
summed SRM chromatograms for the four targeted
rhEPO peptides with their stable-isotope labeled internal
2
4
6
8
10
12
14
Time (min)
0
50
100
0
50
100
0
50
100
0
50
100
RelativeAbundance
0
50
100
0
50
100
1.60
8.51
2.39
9.94
50.21
22.4
7.19
4.96
13.13
9.60
10.28
8.45
11.36 13.42
1.14
5.59
4.73
2.90
6.84
10.06
10.45 12.06
1.32
13.78
5.82 6.84
3.88
2.91
9.66
11.20
12.51
10.85
13.42
6.84
1.32
5.59
3.88
2.91
9.60
12.06
12.34
10.22
3.20
9.31
6.61
1.31
5.59
12.63
12.80
11.83
9.89
4.91
1.77 2.91
5.82 7.25 8.91
T10 AVSGLR
T4 YLLEAK
T14 TITADTFR
T17 VYSNFLR
T11 SLTTLLR
T6 YNFYAWK
2
4
6
8
10
12
14
Time (min)
0
50
100
0
50
100
0
50
100
0
50
100
0
50
100
0
50
100
1.43
2.05 3.19 4.78 5.40 6.88 8.82
12.18
9.67
12.92
9.50
10.07
5.63
9.22
12.86
2.62 4.61
6.77
10.08
10.42
8.88
12.70
2.974.05 5.98
1.83
7.79
11.33
11.73 13.15
5.92
10.19
4.27
3.02
1.71
7.79
12.07
12.30
11.79
7.45
0.97 2.96
5.41
9.11
3.99
12.64
13.04
11.96
10.02
4.56
7.79
3.19
5.98
1.72
T10 AVSGLR
T4 YLLEAK
T14 TITADTFR
T17 VYSNFLR
T11 SLTTLLR
T6 YNFYAWK
1A)
Figure 1: SRM chromatographic traces for each of the targeted peptides for
1A) DPO and 1B) rhEPO enzymatic digest using identical experimental method.
1B)
0
2
4
6
8
10
12
14
16
18
Time (min)
Relative Abundance
0
100
0
100
0
100
0
100
0
100
0
100
0
100
0
100
RT: 7.18
MA: 13527
RT: 7.10
MA: 2368799
RT: 9.27
MA: 10653
RT: 9.26
MA: 2331810
RT: 8.68
MA: 2926
RT: 8.71
MA: 651563
RT: 9.68
MA: 1205
RT: 9.59
MA: 429655
NL: 2.44E3
NL: 2.45E5
NL: 2.64E3
NL: 4.25E5
NL: 6.59E2
NL: 1.22E5
NL: 3.66E2
NL: 8.75E4
YLLEAK
Y(*L)LEAK
SLTTLLR
S(*L)TTLLR
VYSNFLR
VYSN(*F)LR
VYNFAWK
VYN(*F)AWK
T4
T11
T17
T6
Figure 2: SRM responses for four targeted rhEPO peptides and the
corresponding stable isotope labeled peptide. The measured response is for
a total of 500 amol on column for the unlabeled rhEPO and 100 fmol for the
labeled rhEPO peptides.
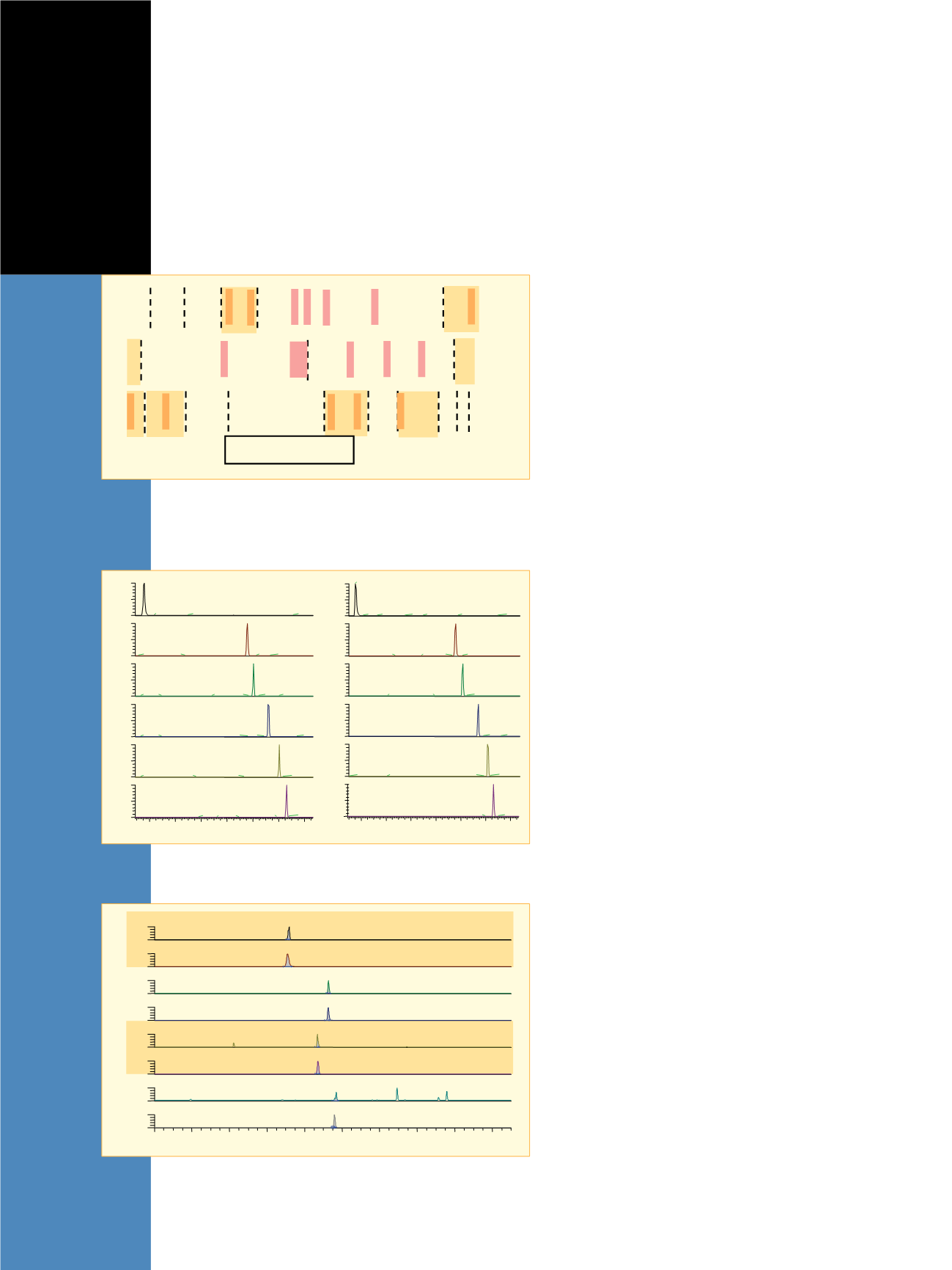
Legend: Protein Sequences
rhEPO
DPO
eEPO
T4
T5
T10
T11
T14
T17
APPRLICDSR VLERYLLEAK EAENI TTGCA EHCSLNEN IT VPDTKVNFYA
APPRLICDSR VLERYLLEAK EAENI TTGCN ETCSLNEN IT VPDTKVNFYA
**PPRLICDSR VLERYILEAR EAENVTMGCA EGCSFGENVT VPDTKVNFYS
WKRMEVGQQA VEVWQGLALL SEAVLRGQAL LVNSSQPWEP LQLHVDKAVS
WKRMEVGQQA VEVQQGLALL SEAVLRGQAL LVNSSQVNET LQLHVDKAVS
WKRMEVGQQA VEVWQGLALL SEAITQGQAL LANSSQPSET LRLGVDKAVS
GLRSLTTLLR ALGAQKEAIS PPDAASAAPL RTITADTFRK LFRVYSNFLR GKLKLYTGEA
GLRSLTTLLR ALGAQKEAIS PPDAASAAPL RTITADTFRK LFRVYSNFLR GKLKLYTGEA
SLRSLTSLLR ALGAQKEAIS PPDAASAAPL RTFAVDTLCK LFR IYSNFLR GKLKLYTGEA
CRTGD
CRTGD
CRR
Scheme 1. Comparison of protein sequences for rhEPO, DPO, and equine
EPO. The dashed lines represent sites of enzymatic cleavages and the red
boxes highlight non-conserved sequence sites between rhEPO/DPO and
equine EPO. The targeted peptides are marked with a gold box.



















