
5
Thermo Scientific Poster Note
•
PN-MSACL-2014-Intelligent-Acquisition-Prakash_E_03/14S
ed correlation coefficient
on spectra will continue
re 2.
based on high IQ
mine targeted
n list:
overy experiments
way determination
ctional groups
sition methods from
es
and product ion
m/z
tive abundance
dows
ve/absolute
across technical or
licates
Results
Highly multiplexed targeted protein quantification requires significant steps of method
refinement prior to implementation. While the determination of proteins is relatively
straightforward based on biology, the selection of peptides as surrogate biomarkers
and corresponding
m/z
values (precursor and product ions) used to uniquely identify
and quantitate the peptide targets becomes challenging. Generally, retention times
and acquisition windows must be determined to maximize instrument cycle time to
achieve robust quantification. To expedite complex experimental method
development, we have created a unique spectral library procedure based on an
analytically rigorous discovery data acquisition scheme. The local spectral library
contains both LC and MS information that can be readily enlisted to build robust
methods requiring few refinement steps.
To first test our methods, a protein mix was spiked in equine plasma (containing
PTRC kit). Spectral library was first built on the neat protein mixture. Experiments
performed on the quadrupole Orbitrap mass spectrometer facilitate unique product
ion collection and detection schemes to not only increase data acquisition, but
perform state-model data acquisition, increasing the ability for quantification. Figure 3
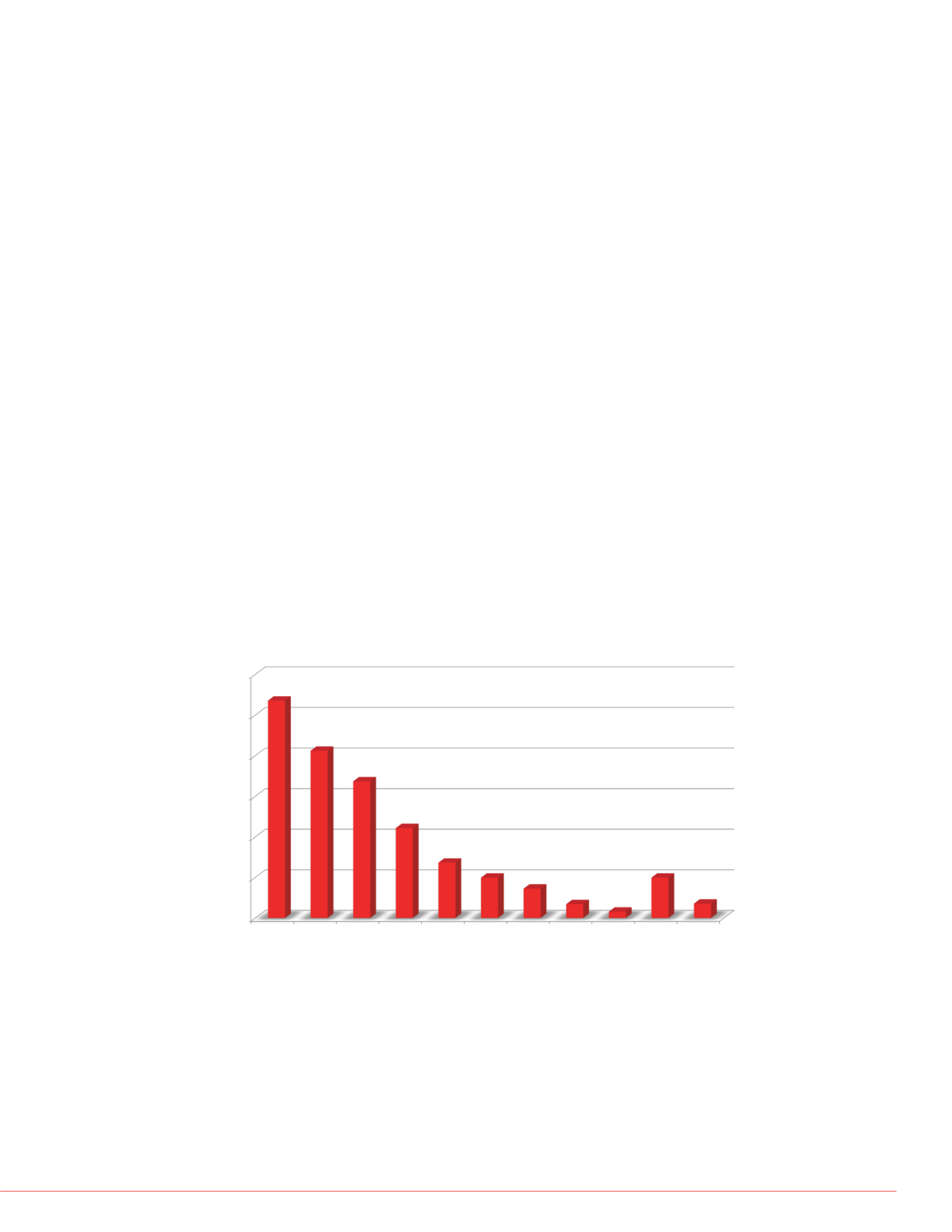
shows the CV distribution for the peptides over four acquisitions (by summing the
area of top eight product ions).
Conclusion
The developments h
3,000 peptides repr
Successful quantific
the ability to change
References
1. Peptide retention
performance liqu
2009
.
All trademarks are the prop
For Research Use Only.
This information is not inte
property rights of others.
K562 Cell Line
2,100 proteins were selected from the K562 cell line and imported into the new
algorithm. The algorithm utilizes the spectral library information to select unique
peptides and create precursor and product ion information used to perform real-time
qualitative and quantitative analysis. In total, 3,800 peptides were chosen and 20-fold
range digest was created.
Figure 4 shows an example where the ratio of 1:10 could not be calculated using the
full-scan MS1 (panel A), but could be calculated in tandem MS/MS scan (panel B,
and zoom-in, panel C).
Product io
C
ntelligent Acquisition
; Tara Schroeder
3
; Lisa Vasicek
6
; Brian Hood
6
; Ryan Bomgarden
4
;
orboys
5
; Claus Jor ensen
5
; T oma Conr ds
6
; Mary Lop z
1
any;
3
Thermo Fisher Scientific Somerset, Somerset, NJ;
4
Thermo Fisher Scientific
at Inova Health, Annandal , VA
ion schemes for
window, target
quisition. Both
to increase the
or “watch list”
Experimental
Spectrum
0
5
10
15
20
25
30
0-2
2-4
4-6
6-8
8-10
10-12
12-15
15-20
20-25
25+
missed
Frequency
CV% Range
FIGURE 3. CV distribution for the initial peptide list.
FIGURE 4. The benefit of MS/MS scan (with higher S/N) compared to full scan. Ratio
of 1:10 could not be calculated in full scan (panel A), but it could be calculated in
tandem MS/MS scan (panel C).
Light Channel
Heavy Channel
A



















