
4
Improving Throughput for Targeted Quantification Methods by Intelligent Acquisition
tion utilizing real-time
over large dynamic ranges.
que to verify putative
These candidates are
ent or derived from a
al interaction. These lists
s spanning several orders
analytical challenges for
pment and throughput. We
ss spectrometry (MS) and
raries for automated method
real-time using novel
t media, collected and mixed
ples were digested and
adrupole-Orbitrap mass
was acquired in two steps
yed unbiased data-
identification as well as
relative retention time,
utions, creating a unique
rgeted protein list was
ata acquisition and
scheme.
re detail. The first step is to
entific™ Pierce™ PRTC
e are interested in. This will
iment. The next is to build a
predictive algorithm or
okup table. The look-up
arge state as well as the
product ion spectral
otopes during the expected
opes surpasses the user-
ciation (HCD) spectrum is
library generating a dot-
p and to check if the
lated correlation coefficient
ct ion spectra will continue
Figure 2.
tion based on high IQ
termine targeted
otein list:
iscovery experiments
athway determination
unctional groups
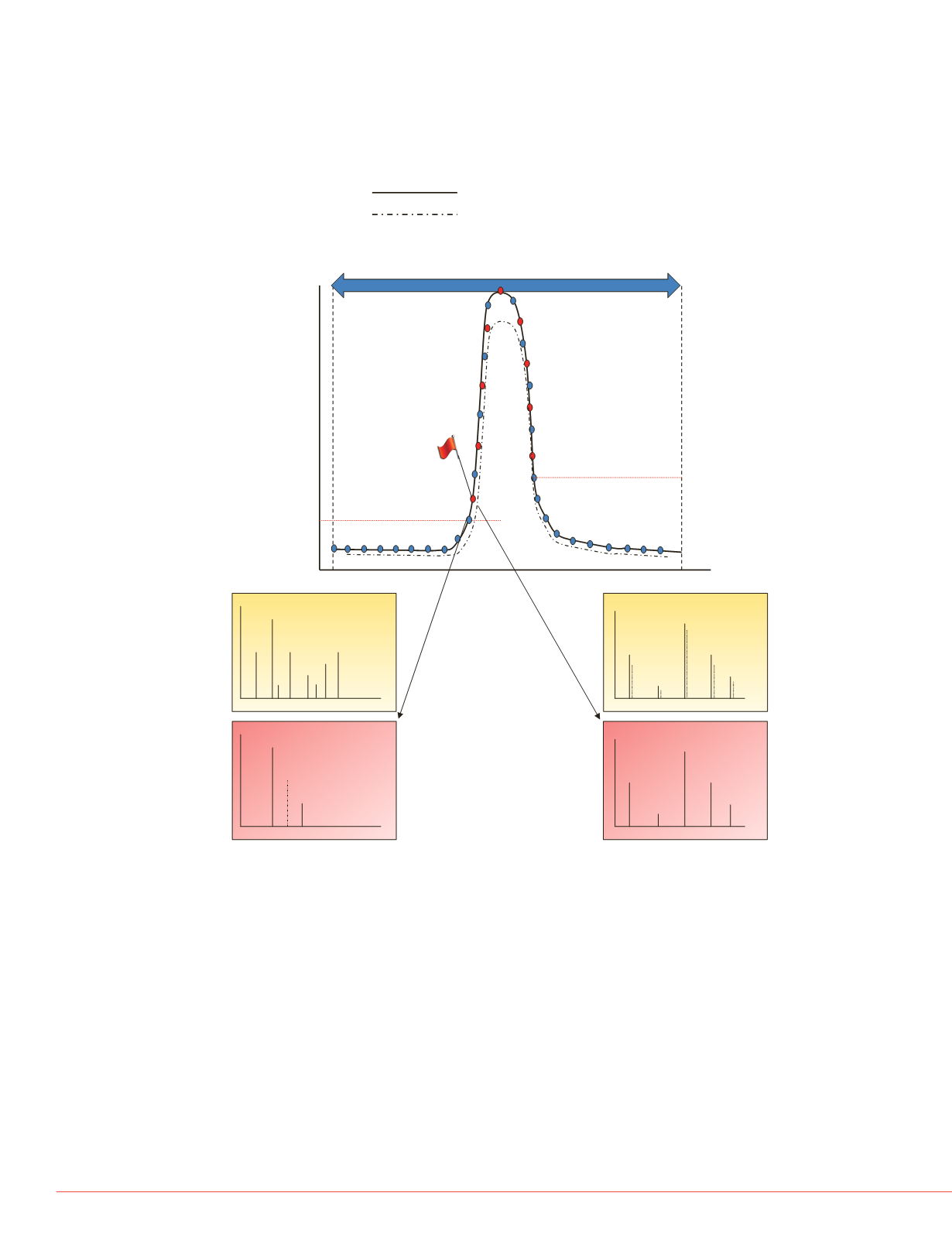
FIGURE 2. Pictorial representation of high IQ data acquisition schemes for
targeted peptide quantification using a targeted scanning window, target
elution identification, and real-time product ion spectral acquisition. Both
precursor and production spectral matching is performed to increase the
analytical selectivity of data acquisition.
*
*
Most intense isotope
2
nd
most intense isotope
Measured Ion Intensity
Retention Time (min)
Start time for “watch list”
Stop time for “watch list”
Triggering
Threshold
1.
Spectral
Library
Experimental
Spectrum
Theoretical
Isotope
Experimental
HR/AM MS
Spectrum
Results
Highly multiplexed targeted protein quantification requires significant steps of method
refinement prior to implementation. While the determination of proteins is relatively
straightforward based on biology, the selection of peptides as surrogate biomarkers
and corresponding
m/z
values (precursor and product ions) used to uniquely identify
and quantitate the peptide targets becomes challenging. Generally, retention times
and acquisition windows must be determined to maximize instrument cycle time to
achieve robust quantification. To expedite complex experimental method
development, we have created a unique spectral library procedure based on an
analytically rigorous discovery data acquisition scheme. The local spectral library
contains both LC and MS information that can be readily enlisted to build robust
methods requiring few refinement steps.
To first test our methods, a protein mix was spiked in equine plasma (containing
PTRC kit). Spectral library was first built on the neat protein mixture. Experiments
performed on the quadrupole Orbitrap mass spectrometer facilitate unique product
ion collection and detection schemes to not only increase data acquisition, but
perform st te-model data acquisition, increasing the ability for quantification. Figure 3
shows the CV distribution for the peptides over four acquisitions (by summing the
0
5
10
15
20
25
30
0-2
2-
Frequency
FIGURE 3. CV d
Conclusion
FIGURE 4. The b
of 1:10 could not
tandem MS/MS s
A
Product
B
C



















