
3
Thermo Scientific Poster Note
•
PN64039 HUPO14_E 04/14S
proteins for the first set of experiments was serotransferrin and
zinc-
α
-2-glycoprotein.
A second set of samples were prepared following the same
approach targeting four different proteins: serotransferrin, zinc-
α
-2-glycoprotein,
α
-1-antitrypsin, and lactotransferrin. Four
different MSIA D.A.R.T. tips were used with each set covalently
bound to the specific Ab. The same set of samples were used
for individual extraction (one tip per sample) followed by
reduction, alkylation, and digestion then LC-MS. The second set
of samples had each vial processed through serial extraction by
different MSIA tip. For example, one sample was first extracted
by anti-serotransferrin, then anti-lactotransferrin etc. and the
extractions were pooled prior to reduction, alkylation, and
digested and ultimately LC-MS.
FIGURE 3. Comparative
result of extraction step
peptide vs. An N-linked
consistent across each
extracted. The two pep
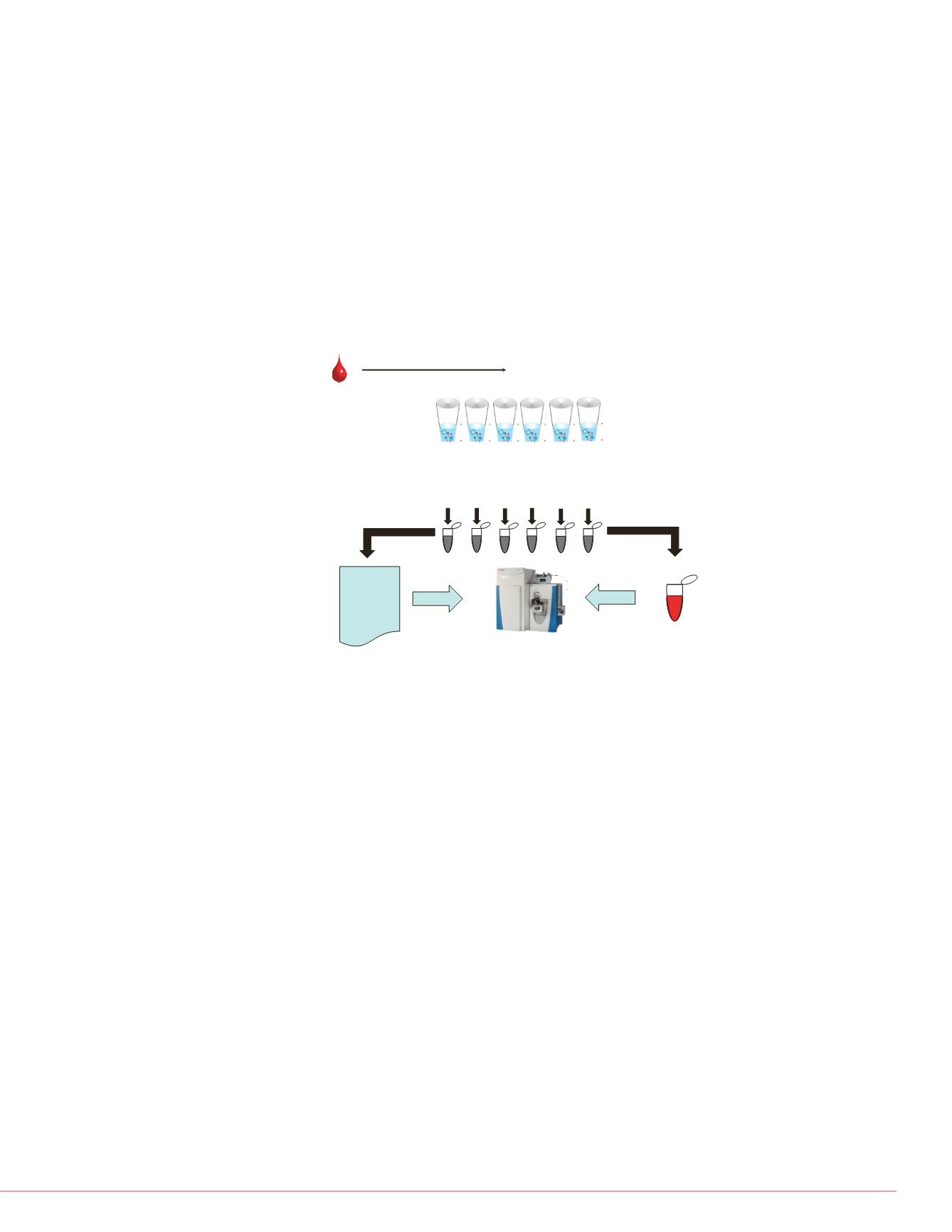
Sample extraction and elution (2 hour)
#1
#3 #4 #5 #6
#2
Same analytical
sample for all
extractions
Acquisition
Queue
#1
#2
#3
#4
….
LC-MS
Single
Pooled
LC-MS
Reduction
Alkylation
Digestion
Reduction
Alkylation
Digestion
FIGURE 1. Strategy for testing the effects of serial dilution
on sample disruption. Two sample preparation types were
individual extraction and analysis vs. pooling samples.
HSTIFENLANK
0
0.5
1
1.5
2
2.5
3
3.5
4
4.5
0
5
10
Peptide AUC Ratios [Sample 2: Sample x]
Serotransferr
FIGURE 4. Comparative
serotransferrin peptides
from the second MSIA e
extractions.
ucibility of serial protein extraction
ual or multiple protein extraction.
S analysis of MSIA D.A.R.T serial
ple using one Ab for a single
er D.A.R.T tip).
1
The relative
used to determine depletion and
ilution.
ractions increased targeted peptide
gnificantly increasing background
bundance resulting from pooling
protein sequence coverage and
lycopeptides and corresponding
using LC-MS provides significant
otein sequence variations,
extend the sensitivity,
ed on antibody (Ab) capture has
fits of Ab capture for LC-MS
protein amounts while
und matrix effects facilitating faster
uence coverage. Performing Ab
metry ImmunoAffinity (MSIA)
ample. This facilitates serial
mes yet facilitating multiplexed
Results
Liquid Chromatography (or more generically Separations)
LC separation was performed using a Thermo Fisher Scientific™
Hypersil Gold™ 100 x 1 mm column with 1.9 µm particle size and
a binary solvent system comprised of A) 0.1% formic acid and B)
0.1% formic acid in MeCN. A linear gradient of 5-32% B was
performed over 15 minutes prior to column washing and re-
equilibration.
Mass Spectrometry
All experiments were performed on a Thermo Scientific™ Q
Exactive™ mass spectrometer operated in data
dependent/dynamic exclusion mode using a Top 10 acquisition
scheme. Full scan MS spectra were acquired using a resolution
setting of 70k and all HCD product ion spectra were acquired
using 15k.
Data Analysis
Initial unbiased data searching was performed using Thermo
Scientific™ Proteome Discoverer™ 1.4 to Thermo Scientific™
Pinpoint 1.4 to search N- and O-linked glycopeptides through the
Screening Tool. To total list of peptides (modified and
unmodified) were used to performed relative quantitation across
all samples. Qualitative assessment was based on retention time
overlap with spectral library values, accurate mass (10 ppm
tolerance), and isotopic distribution. Negative controls were
analyzed by evaluating precursor ion intensities for sample RAW
files not expected to contain the targeted protein.
ltiplexed Protein Quantitation Using Serial Immunoaffinity Ex
C-MS
n,
1
Amol Prakash,
1
Gregory Byram,
1
Maryann Vogelsang,
1
Gouri Vadali,
1
Bryan Krastin
1
Thermo Fisher Scientific BRIMS, Cambridge, MA
FIGURE 6. Comparative t
different sample preparati
histograms report the nor
MSIA extraction per plas
serial MSIA extraction of t
“Serial”). The numbers b
represents the degree of
MS analysis. For exampl
where the alpha-1-antitry
other targeted protein ext
0.0
2
dHex1Hex5HexNAc5NeuAc1
dHex1Hex4HexNAc4NeuAc
dHex1Hex6HexNAc2
Hex6HexNAc5NeuAc3
Hex6HexNAc5NeuAc2
Hex6HexNAc5NeuAc1
Hex5HexNAc4NeuAc2
Hex5HexNAc4NeuAc1
Hex4HexNAc3NeuAc1
Hex6HexNAc6
Hex4HexNAc4
peptide
N-linked Glycan
FIGURE 5. Reproducibilit
comparative AUC ratios f
C*GLVPLAENYN*K.S as



















